| 1 |
|
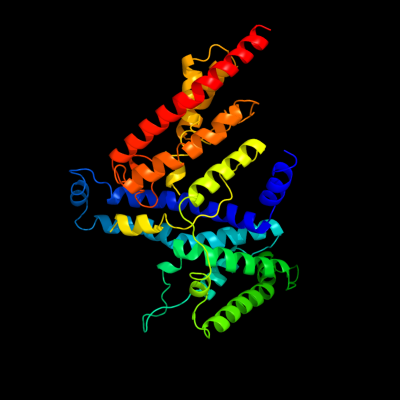
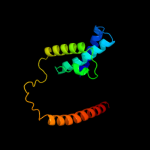
PDB 3qnq chain D
Region: 20 - 414
Aligned: 379
Modelled: 379
Confidence: 100.0%
Identity: 14%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 2 |
|
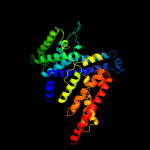
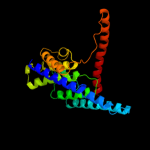
PDB 2voy chain B
Region: 20 - 58
Aligned: 39
Modelled: 39
Confidence: 30.1%
Identity: 15%
PDB header:hydrolase
Chain: B: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 3 |
|
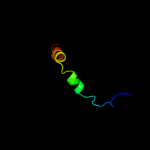
PDB 1u7g chain A
Region: 337 - 406
Aligned: 70
Modelled: 70
Confidence: 27.9%
Identity: 16%
Fold: Ammonium transporter
Superfamily: Ammonium transporter
Family: Ammonium transporter
Phyre2
| 4 |
|
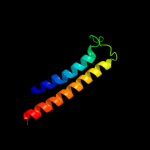
PDB 3dtu chain B domain 2
Region: 378 - 409
Aligned: 32
Modelled: 32
Confidence: 9.4%
Identity: 13%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 5 |
|
PDB 1ffy chain A domain 1
Region: 17 - 51
Aligned: 35
Modelled: 35
Confidence: 9.0%
Identity: 3%
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
Superfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
Family: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
Phyre2
| 6 |
|
PDB 1fft chain B domain 2
Region: 379 - 409
Aligned: 31
Modelled: 31
Confidence: 8.1%
Identity: 10%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 7 |
|
PDB 2ket chain A
Region: 32 - 50
Aligned: 19
Modelled: 19
Confidence: 6.3%
Identity: 16%
PDB header:antibiotic
Chain: A: PDB Molecule:cathelicidin-6;
PDBTitle: solution structure of bmap-27
Phyre2
| 8 |
|
PDB 3ehb chain B domain 2
Region: 378 - 409
Aligned: 32
Modelled: 32
Confidence: 6.2%
Identity: 13%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 9 |
|
PDB 3nd0 chain A
Region: 284 - 409
Aligned: 126
Modelled: 126
Confidence: 6.2%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:sll0855 protein;
PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
Phyre2
| 10 |
|
PDB 2ht2 chain B
Region: 164 - 409
Aligned: 237
Modelled: 240
Confidence: 6.2%
Identity: 10%
PDB header:membrane protein
Chain: B: PDB Molecule:h(+)/cl(-) exchange transporter clca;
PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
Phyre2