1 c1vzmB_
19.7
100
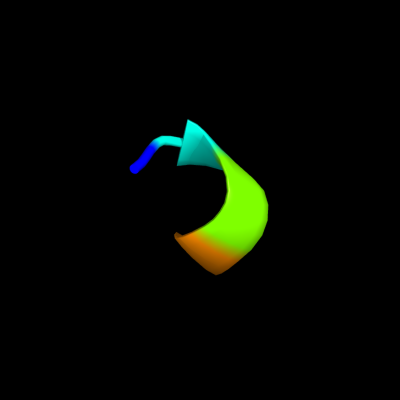
PDB header: calcium-binding proteinChain: B: PDB Molecule: osteocalcin;PDBTitle: osteocalcin from fish argyrosomus regius
2 d2cqka1
17.9
29
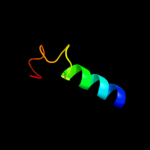
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: La domain3 d1zh5a1
15.4
24
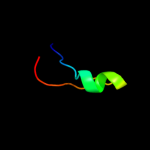
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: La domain4 c3pf6C_
13.0
45
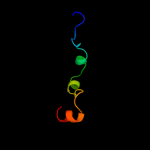
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein pp-luz7_gp033;PDBTitle: the structure of uncharacterized protein pp-luz7_gp033 from2 pseudomonas phage luz7.
5 d2k0bx1
12.7
67
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain6 d2nrqa1
12.7
48
Fold: RL5-likeSuperfamily: RL5-likeFamily: SSO1042-like7 d1s29a_
12.0
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: La domain8 c2qnlA_
11.7
32
PDB header: signaling proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative dna damage-inducible protein2 (chu_0679) from cytophaga hutchinsonii atcc 33406 at 1.50 a3 resolution
9 d1rzhh1
10.5
14
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: Photosynthetic reaction centre, H-chain, cytoplasmic domain10 c2i6hA_
10.3
46
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein atu0120;PDBTitle: structure of protein of unknown function atu0120 from agrobacterium2 tumefaciens
11 d2i6ha1
10.3
46
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Atu0120-like12 d2r48a1
10.0
17
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like13 d2r4qa1
9.7
33
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like14 c2k1aA_
8.7
46
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
15 d1hf8a1
8.7
20
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: Phosphoinositide-binding clathrin adaptor, domain 216 d1dfma_
8.4
32
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease BglII17 c2kncA_
7.8
46
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
18 d2jdid1
7.6
30
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase19 c2i5nH_
7.5
32
PDB header: photosynthesisChain: H: PDB Molecule: reaction center protein h chain;PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
20 d2gica1
7.4
38
Fold: Rhabdovirus nucleoprotein-likeSuperfamily: Rhabdovirus nucleoprotein-likeFamily: Rhabdovirus nucleocapsid protein21 c3izxE_
not modelled
7.2
23
PDB header: virusChain: E: PDB Molecule: viral structural protein 5;PDBTitle: 3.1 angstrom cryoem structure of cytoplasmic polyhedrosis virus
22 c1eysH_
not modelled
7.2
25
PDB header: electron transportChain: H: PDB Molecule: photosynthetic reaction center;PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
23 d2ogka1
not modelled
7.2
29
Fold: RL5-likeSuperfamily: RL5-likeFamily: SSO1042-like24 d1hx8a1
not modelled
7.0
15
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: Phosphoinositide-binding clathrin adaptor, domain 225 c3e5aB_
not modelled
6.8
69
PDB header: transferaseChain: B: PDB Molecule: targeting protein for xklp2;PDBTitle: crystal structure of aurora a in complex with vx-680 and tpx2
26 c2ks1B_
not modelled
6.8
56
PDB header: transferaseChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
27 d1lghb_
not modelled
6.7
60
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits28 c3lpzA_
not modelled
6.4
16
PDB header: protein transportChain: A: PDB Molecule: get4 (yor164c homolog);PDBTitle: crystal structure of c. therm. get4
29 c2xvoB_
not modelled
5.7
47
PDB header: structural genomicsChain: B: PDB Molecule: sso1725;PDBTitle: sso1725, a protein involved in the crispr/cas pathway
30 d1e6yb2
not modelled
5.7
31
Fold: Ferredoxin-likeSuperfamily: Methyl-coenzyme M reductase subunitsFamily: Methyl-coenzyme M reductase alpha and beta chain N-terminal domain31 c1ponB_
not modelled
5.7
50
PDB header: calcium-binding proteinChain: B: PDB Molecule: troponin c;PDBTitle: site iii-site iv troponin c heterodimer, nmr
32 c3pryA_
not modelled
5.7
70
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hsp 90-beta;PDBTitle: crystal structure of the middle domain of human hsp90-beta refined at2 2.3 a resolution
33 c2pqlA_
not modelled
5.7
40
PDB header: transport proteinChain: A: PDB Molecule: d7r4 protein;PDBTitle: crystal structure of anopheles gambiae d7r4-tryptamine complex
34 d1qkya_
not modelled
5.6
80
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins35 c2wnyB_
not modelled
5.6
43
PDB header: unknown functionChain: B: PDB Molecule: conserved protein mth689;PDBTitle: structure of mth689, a duf54 protein from methanothermobacter2 thermautotrophicus
36 d1h5wa_
not modelled
5.6
18
Fold: Upper collar protein gp10 (connector protein)Superfamily: Upper collar protein gp10 (connector protein)Family: Upper collar protein gp10 (connector protein)37 c2nr1A_
not modelled
5.4
57
PDB header: receptorChain: A: PDB Molecule: nr1 m2;PDBTitle: transmembrane segment 2 of nmda receptor nr1, nmr, 102 structures
38 c1y6zA_
not modelled
5.4
60
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: heat shock protein, putative;PDBTitle: middle domain of plasmodium falciparum putative heat shock protein2 pf14_0417
39 d2v7qi1
not modelled
5.4
44
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Epsilon subunit of mitochondrial F1F0-ATP synthaseFamily: Epsilon subunit of mitochondrial F1F0-ATP synthase40 d1q90g_
not modelled
5.3
56
Fold: Single transmembrane helixSuperfamily: PetG subunit of the cytochrome b6f complexFamily: PetG subunit of the cytochrome b6f complex41 c1q90G_
not modelled
5.3
56
PDB header: photosynthesisChain: G: PDB Molecule: cytochrome b6f complex subunit petg;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
42 d3d85d2
not modelled
5.1
38
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Fibronectin type IIIFamily: Fibronectin type III