| 1 |
|
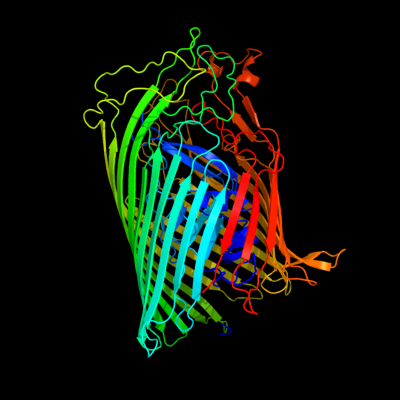
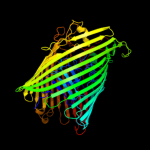
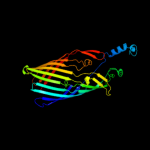
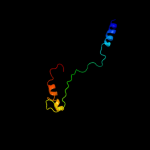
PDB 2iah chain A
Region: 18 - 790
Aligned: 664
Modelled: 697
Confidence: 100.0%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:ferripyoverdine receptor;
PDBTitle: crystal structure of the ferripyoverdine receptor of the outer2 membrane of pseudomonas aeruginosa bound to ferripyoverdine.
Phyre2
| 2 |
|
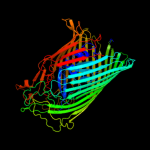
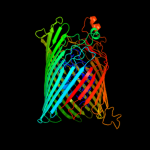
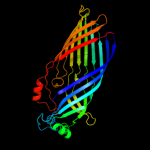
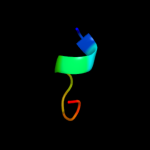
PDB 1by5 chain A
Region: 19 - 790
Aligned: 685
Modelled: 699
Confidence: 100.0%
Identity: 11%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Ligand-gated protein channel
Phyre2
| 3 |
|
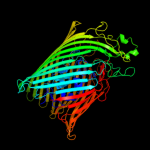
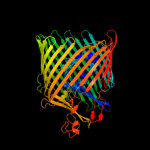
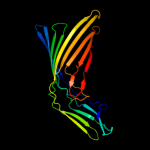
PDB 2grx chain B
Region: 20 - 790
Aligned: 678
Modelled: 678
Confidence: 100.0%
Identity: 12%
PDB header:metal transport
Chain: B: PDB Molecule:ferrichrome-iron receptor;
PDBTitle: crystal structure of tonb in complex with fhua, e. coli2 outer membrane receptor for ferrichrome
Phyre2
| 4 |
|
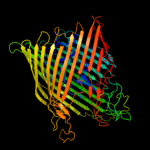
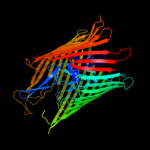
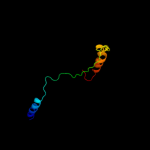
PDB 1xkh chain C
Region: 14 - 790
Aligned: 673
Modelled: 662
Confidence: 100.0%
Identity: 11%
PDB header:membrane protein
Chain: C: PDB Molecule:ferripyoverdine receptor;
PDBTitle: pyoverdine outer membrane receptor fpva from pseudomonas aeruginosa2 pao1 bound to pyoverdine
Phyre2
| 5 |
|
PDB 3fhh chain A
Region: 31 - 790
Aligned: 607
Modelled: 626
Confidence: 100.0%
Identity: 18%
PDB header:membrane protein
Chain: A: PDB Molecule:outer membrane heme receptor shua;
PDBTitle: crystal structure of the heme/hemoglobin outer membrane2 transporter shua from shigella dysenteriae
Phyre2
| 6 |
|
PDB 1xkw chain A
Region: 24 - 790
Aligned: 644
Modelled: 675
Confidence: 100.0%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:fe(iii)-pyochelin receptor;
PDBTitle: pyochelin outer membrane receptor fpta from pseudomonas2 aeruginosa
Phyre2
| 7 |
|
PDB 3qlb chain A
Region: 21 - 790
Aligned: 657
Modelled: 676
Confidence: 100.0%
Identity: 11%
PDB header:metal transport
Chain: A: PDB Molecule:enantio-pyochelin receptor;
PDBTitle: enantiopyochelin outer membrane tonb-dependent transporter from2 pseudomonas fluorescens bound to the ferri-enantiopyochelin
Phyre2
| 8 |
|
PDB 3csl chain B
Region: 39 - 790
Aligned: 689
Modelled: 708
Confidence: 100.0%
Identity: 13%
PDB header:membrane protein/heme binding protein
Chain: B: PDB Molecule:hasr protein;
PDBTitle: structure of the serratia marcescens hemophore receptor hasr in2 complex with its hemophore hasa and heme
Phyre2
| 9 |
|
PDB 1fep chain A
Region: 31 - 790
Aligned: 660
Modelled: 689
Confidence: 100.0%
Identity: 13%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Ligand-gated protein channel
Phyre2
| 10 |
|
PDB 1kmo chain A
Region: 32 - 790
Aligned: 646
Modelled: 672
Confidence: 100.0%
Identity: 14%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Ligand-gated protein channel
Phyre2
| 11 |
|
PDB 1po3 chain A
Region: 38 - 790
Aligned: 635
Modelled: 658
Confidence: 100.0%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:iron(iii) dicitrate transport protein feca
PDBTitle: crystal structure of ferric citrate transporter feca in2 complex with ferric citrate
Phyre2
| 12 |
|
PDB 2guf chain A domain 1
Region: 31 - 790
Aligned: 540
Modelled: 602
Confidence: 100.0%
Identity: 18%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Ligand-gated protein channel
Phyre2
| 13 |
|
PDB 2hdi chain A
Region: 31 - 790
Aligned: 570
Modelled: 599
Confidence: 100.0%
Identity: 15%
PDB header:protein transport,antimicrobial protein
Chain: A: PDB Molecule:colicin i receptor;
PDBTitle: crystal structure of the colicin i receptor cir from e.coli in complex2 with receptor binding domain of colicin ia.
Phyre2
| 14 |
|
PDB 3efm chain A
Region: 18 - 769
Aligned: 552
Modelled: 573
Confidence: 100.0%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:ferric alcaligin siderophore receptor;
PDBTitle: structure of the alcaligin outer membrane recepteur faua from2 bordetella pertussis
Phyre2
| 15 |
|
PDB 1t16 chain A
Region: 485 - 790
Aligned: 277
Modelled: 286
Confidence: 99.2%
Identity: 8%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Outer membrane protein transport protein
Phyre2
| 16 |
|
PDB 3dwo chain X
Region: 377 - 742
Aligned: 309
Modelled: 324
Confidence: 94.9%
Identity: 9%
PDB header:membrane protein
Chain: X: PDB Molecule:probable outer membrane protein;
PDBTitle: crystal structure of a pseudomonas aeruginosa fadl homologue
Phyre2
| 17 |
|
PDB 3brz chain A
Region: 469 - 742
Aligned: 213
Modelled: 223
Confidence: 90.0%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:todx;
PDBTitle: crystal structure of the pseudomonas putida toluene2 transporter todx
Phyre2
| 18 |
|
PDB 3k07 chain A
Region: 4 - 75
Aligned: 63
Modelled: 70
Confidence: 29.9%
Identity: 6%
PDB header:transport protein
Chain: A: PDB Molecule:cation efflux system protein cusa;
PDBTitle: crystal structure of cusa
Phyre2
| 19 |
|
PDB 1oy8 chain A
Region: 4 - 74
Aligned: 62
Modelled: 71
Confidence: 24.8%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:acriflavine resistance protein b;
PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
Phyre2
| 20 |
|
PDB 2a1j chain A domain 1
Region: 66 - 75
Aligned: 10
Modelled: 10
Confidence: 20.0%
Identity: 40%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Hef domain-like
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|