| 1 |
|
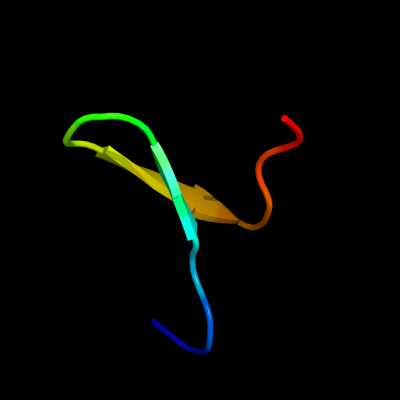
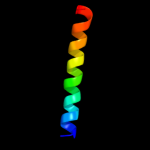
PDB 2k89 chain A
Region: 59 - 75
Aligned: 17
Modelled: 17
Confidence: 38.7%
Identity: 18%
PDB header:protein binding
Chain: A: PDB Molecule:phospholipase a-2-activating protein;
PDBTitle: solution structure of a novel ubiquitin-binding domain from2 human plaa (pfuc, gly76-pro77 cis isomer)
Phyre2
| 2 |
|
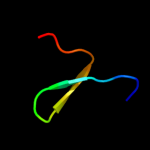
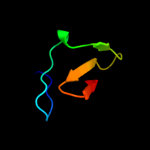
PDB 3pst chain A
Region: 61 - 75
Aligned: 15
Modelled: 15
Confidence: 30.3%
Identity: 33%
PDB header:nuclear protein
Chain: A: PDB Molecule:protein doa1;
PDBTitle: crystal structure of pul and pfu(mutate) domain
Phyre2
| 3 |
|
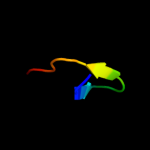
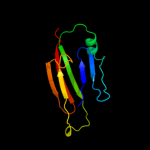
PDB 3l3f chain X
Region: 61 - 75
Aligned: 15
Modelled: 15
Confidence: 27.8%
Identity: 33%
PDB header:protein binding
Chain: X: PDB Molecule:protein doa1;
PDBTitle: crystal structure of a pfu-pul domain pair of saccharomyces cerevisiae2 doa1/ufd3
Phyre2
| 4 |
|
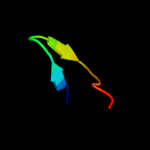
PDB 3c0n chain A domain 1
Region: 47 - 79
Aligned: 25
Modelled: 33
Confidence: 25.9%
Identity: 32%
Fold: C-type lectin-like
Superfamily: C-type lectin-like
Family: Aerolysin/Pertussis toxin (APT) domain
Phyre2
| 5 |
|
PDB 2e76 chain D
Region: 3 - 29
Aligned: 27
Modelled: 27
Confidence: 18.7%
Identity: 7%
PDB header:photosynthesis
Chain: D: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
Phyre2
| 6 |
|
PDB 1oe1 chain A domain 2
Region: 58 - 78
Aligned: 21
Modelled: 21
Confidence: 14.1%
Identity: 19%
Fold: Cupredoxin-like
Superfamily: Cupredoxins
Family: Multidomain cupredoxins
Phyre2
| 7 |
|
PDB 2cs7 chain A domain 1
Region: 39 - 73
Aligned: 35
Modelled: 35
Confidence: 11.4%
Identity: 23%
Fold: IL8-like
Superfamily: PhtA domain-like
Family: PhtA domain-like
Phyre2
| 8 |
|
PDB 2pq4 chain B
Region: 1 - 18
Aligned: 18
Modelled: 18
Confidence: 10.8%
Identity: 39%
PDB header:chaperone/oxidoreductase
Chain: B: PDB Molecule:periplasmic nitrate reductase precursor;
PDBTitle: nmr solution structure of napd in complex with napa1-352 signal peptide
Phyre2
| 9 |
|
PDB 1pgl chain 1 domain 1
Region: 50 - 152
Aligned: 102
Modelled: 103
Confidence: 7.0%
Identity: 10%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: Positive stranded ssRNA viruses
Family: Comoviridae-like VP
Phyre2
| 10 |
|
PDB 1hq0 chain A
Region: 21 - 26
Aligned: 6
Modelled: 6
Confidence: 5.3%
Identity: 67%
Fold: CNF1/YfiH-like putative cysteine hydrolases
Superfamily: CNF1/YfiH-like putative cysteine hydrolases
Family: Type 1 cytotoxic necrotizing factor, catalytic domain
Phyre2