| 1 |
|
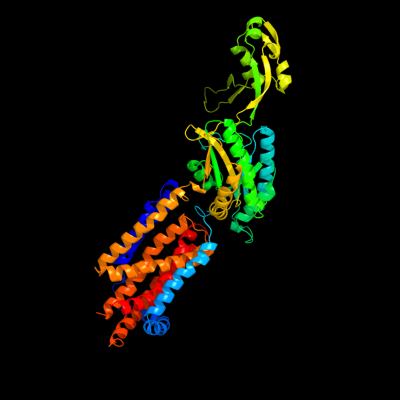
PDB 3k07 chain A
Region: 13 - 605
Aligned: 563
Modelled: 593
Confidence: 100.0%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:cation efflux system protein cusa;
PDBTitle: crystal structure of cusa
Phyre2
| 2 |
|
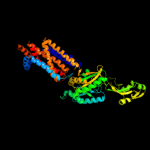
PDB 1oy8 chain A
Region: 13 - 611
Aligned: 553
Modelled: 584
Confidence: 100.0%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:acriflavine resistance protein b;
PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
Phyre2
| 3 |
|
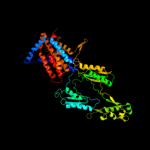
PDB 3aqp chain B
Region: 96 - 614
Aligned: 419
Modelled: 420
Confidence: 100.0%
Identity: 36%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 4 |
|
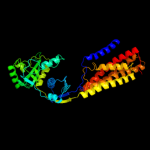
PDB 1iwg chain A domain 8
Region: 411 - 611
Aligned: 200
Modelled: 201
Confidence: 100.0%
Identity: 17%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 5 |
|
PDB 1iwg chain A domain 7
Region: 410 - 606
Aligned: 194
Modelled: 197
Confidence: 99.9%
Identity: 15%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 6 |
|
PDB 3aqo chain D
Region: 216 - 441
Aligned: 207
Modelled: 207
Confidence: 99.7%
Identity: 34%
PDB header:membrane protein
Chain: D: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: structure and function of a membrane component secdf that enhances2 protein export
Phyre2
| 7 |
|
PDB 1iwg chain A domain 6
Region: 292 - 382
Aligned: 88
Modelled: 91
Confidence: 98.7%
Identity: 6%
Fold: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains
Superfamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains
Family: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains
Phyre2
| 8 |
|
PDB 1iwg chain A domain 5
Region: 293 - 382
Aligned: 89
Modelled: 90
Confidence: 95.8%
Identity: 8%
Fold: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains
Superfamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains
Family: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains
Phyre2
| 9 |
|
PDB 1iwg chain A domain 4
Region: 371 - 437
Aligned: 59
Modelled: 67
Confidence: 95.2%
Identity: 10%
Fold: Ferredoxin-like
Superfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains
Family: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains
Phyre2
| 10 |
|
PDB 1iwg chain A domain 2
Region: 340 - 440
Aligned: 94
Modelled: 101
Confidence: 94.5%
Identity: 6%
Fold: Ferredoxin-like
Superfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains
Family: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains
Phyre2
| 11 |
|
PDB 1iwg chain A domain 1
Region: 139 - 207
Aligned: 69
Modelled: 69
Confidence: 59.5%
Identity: 7%
Fold: Ferredoxin-like
Superfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains
Family: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains
Phyre2
| 12 |
|
PDB 1cii chain A
Region: 488 - 523
Aligned: 36
Modelled: 36
Confidence: 41.1%
Identity: 19%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 13 |
|
PDB 2bbj chain B
Region: 517 - 609
Aligned: 86
Modelled: 93
Confidence: 34.8%
Identity: 8%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 14 |
|
PDB 2r6g chain F domain 1
Region: 8 - 34
Aligned: 26
Modelled: 27
Confidence: 15.4%
Identity: 19%
Fold: MalF N-terminal region-like
Superfamily: MalF N-terminal region-like
Family: MalF N-terminal region-like
Phyre2
| 15 |
|
PDB 2yvx chain D
Region: 476 - 615
Aligned: 137
Modelled: 140
Confidence: 14.7%
Identity: 13%
PDB header:transport protein
Chain: D: PDB Molecule:mg2+ transporter mgte;
PDBTitle: crystal structure of magnesium transporter mgte
Phyre2
| 16 |
|
PDB 2qts chain A
Region: 487 - 511
Aligned: 23
Modelled: 25
Confidence: 10.9%
Identity: 30%
PDB header:membrane protein
Chain: A: PDB Molecule:acid-sensing ion channel;
PDBTitle: structure of an acid-sensing ion channel 1 at 1.9 a resolution and low2 ph
Phyre2
| 17 |
|
PDB 2iub chain A domain 2
Region: 564 - 609
Aligned: 35
Modelled: 46
Confidence: 8.3%
Identity: 11%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 18 |
|
PDB 1pby chain C
Region: 1 - 8
Aligned: 8
Modelled: 8
Confidence: 7.0%
Identity: 63%
Fold: Non-globular all-alpha subunits of globular proteins
Superfamily: Quinohemoprotein amine dehydrogenase C chain
Family: Quinohemoprotein amine dehydrogenase C chain
Phyre2
| 19 |
|
PDB 1xme chain B domain 2
Region: 449 - 471
Aligned: 23
Modelled: 23
Confidence: 6.8%
Identity: 22%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 20 |
|
PDB 1jmx chain G
Region: 1 - 8
Aligned: 8
Modelled: 8
Confidence: 6.3%
Identity: 38%
Fold: Non-globular all-alpha subunits of globular proteins
Superfamily: Quinohemoprotein amine dehydrogenase C chain
Family: Quinohemoprotein amine dehydrogenase C chain
Phyre2
| 21 |
|
| 22 |
|