| 1 |
|
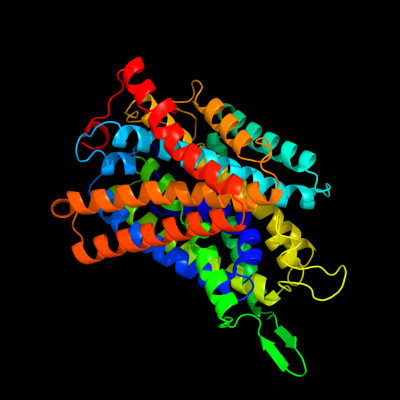
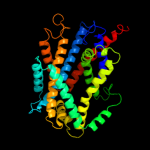
PDB 3gia chain A
Region: 14 - 482
Aligned: 429
Modelled: 454
Confidence: 100.0%
Identity: 19%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
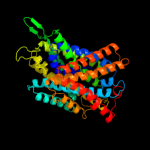
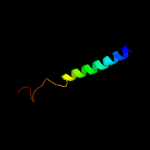
PDB 3lrc chain C
Region: 14 - 478
Aligned: 408
Modelled: 408
Confidence: 100.0%
Identity: 18%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
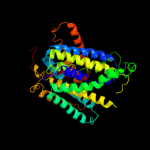
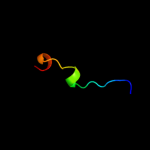
PDB 2jln chain A
Region: 3 - 487
Aligned: 452
Modelled: 474
Confidence: 100.0%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
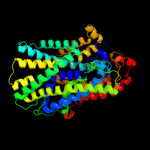
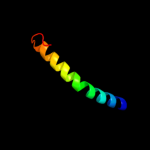
PDB 2xq2 chain A
Region: 15 - 489
Aligned: 449
Modelled: 449
Confidence: 99.5%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 5 |
|
PDB 3dh4 chain A
Region: 15 - 489
Aligned: 447
Modelled: 456
Confidence: 99.3%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 18 - 474
Aligned: 449
Modelled: 449
Confidence: 98.2%
Identity: 14%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 10 - 413
Aligned: 390
Modelled: 386
Confidence: 97.3%
Identity: 9%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 52 - 402
Aligned: 343
Modelled: 351
Confidence: 92.4%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 2knc chain A
Region: 450 - 488
Aligned: 39
Modelled: 39
Confidence: 55.5%
Identity: 15%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 10 |
|
PDB 1ujl chain A
Region: 7 - 28
Aligned: 22
Modelled: 22
Confidence: 17.7%
Identity: 23%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h
PDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
Phyre2
| 11 |
|
PDB 2p8z chain S
Region: 471 - 489
Aligned: 19
Modelled: 19
Confidence: 16.5%
Identity: 5%
PDB header:translation
Chain: S: PDB Molecule:elongation factor tu-b;
PDBTitle: fitted structure of adpr-eef2 in the 80s:adpr-2 eef2:gdpnp:sordarin cryo-em reconstruction
Phyre2
| 12 |
|
PDB 3qnq chain D
Region: 453 - 489
Aligned: 37
Modelled: 37
Confidence: 15.4%
Identity: 5%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 13 |
|
PDB 3b8e chain C
Region: 383 - 477
Aligned: 95
Modelled: 95
Confidence: 12.6%
Identity: 8%
PDB header:hydrolase/transport protein
Chain: C: PDB Molecule:sodium/potassium-transporting atpase subunit
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 14 |
|
PDB 1fft chain B domain 2
Region: 368 - 443
Aligned: 76
Modelled: 76
Confidence: 7.6%
Identity: 9%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 15 |
|
PDB 2zxe chain A
Region: 371 - 474
Aligned: 104
Modelled: 104
Confidence: 6.8%
Identity: 7%
PDB header:hydrolase/transport protein
Chain: A: PDB Molecule:na, k-atpase alpha subunit;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
Phyre2
| 16 |
|
PDB 2bbj chain B
Region: 418 - 474
Aligned: 51
Modelled: 57
Confidence: 5.9%
Identity: 12%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 17 |
|
PDB 1m57 chain H
Region: 368 - 437
Aligned: 70
Modelled: 70
Confidence: 5.5%
Identity: 11%
PDB header:oxidoreductase
Chain: H: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
Phyre2