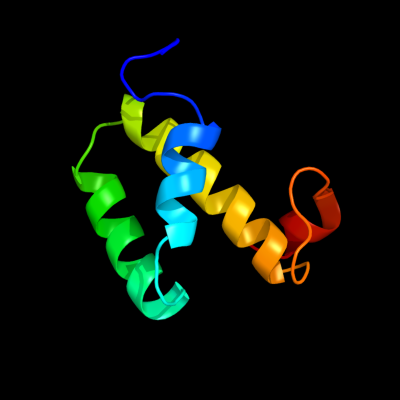
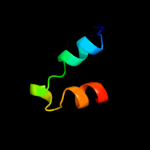
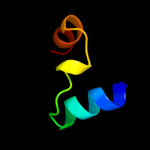
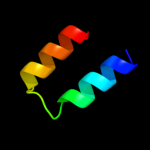
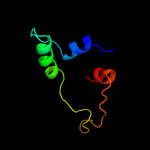
1 d1jw2a_
100.0
100
Fold: Open three-helical up-and-down bundleSuperfamily: Hemolysin expression modulating protein HHAFamily: Hemolysin expression modulating protein HHA2 c2jqtA_
99.6
40
PDB header: protein bindingChain: A: PDB Molecule: h-ns/stpa-binding protein 2;PDBTitle: structure of the bacterial replication origin-associated2 protein cnu
3 d1t1ua1
25.8
7
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase4 d1xl7a1
25.3
14
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase5 c2jpnA_
25.0
27
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent dna helicase uvsw;PDBTitle: solution structure of t4 bacteriophage helicase uvsw.1
6 d1trra_
22.8
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: Trp repressor, TrpR7 d1nm8a1
21.7
18
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase8 c2fyoA_
20.8
21
PDB header: transferaseChain: A: PDB Molecule: carnitine o-palmitoyltransferase ii,PDBTitle: crystal structure of rat carnitine palmitoyltransferase 22 in space group p43212
9 d1ndba1
18.3
18
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase10 c1q6xA_
17.9
7
PDB header: transferaseChain: A: PDB Molecule: choline o-acetyltransferase;PDBTitle: crystal structure of rat choline acetyltransferase
11 d1jhga_
17.9
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: Trp repressor, TrpR12 c2h4tB_
16.6
21
PDB header: transferaseChain: B: PDB Molecule: carnitine o-palmitoyltransferase ii,PDBTitle: crystal structure of rat carnitine palmitoyltransferase ii
13 c3cm8A_
15.4
39
PDB header: rna binding protein/transferaseChain: A: PDB Molecule: polymerase acidic protein;PDBTitle: a rna polymerase subunit structure from virus
14 c1xl8B_
15.4
14
PDB header: transferaseChain: B: PDB Molecule: peroxisomal carnitine o-octanoyltransferase;PDBTitle: crystal structure of mouse carnitine octanoyltransferase in2 complex with octanoylcarnitine
15 d1i27a_
14.0
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal domain of the rap74 subunit of TFIIF16 d1ej2a_
13.7
25
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Adenylyltransferase17 c1t7qA_
13.6
18
PDB header: transferaseChain: A: PDB Molecule: carnitine acetyltransferase;PDBTitle: crystal structure of the f565a mutant of murine carnitine2 acetyltransferase in complex with carnitine and coa
18 d2bsza1
13.2
27
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase19 c2fy2A_
12.8
11
PDB header: transferaseChain: A: PDB Molecule: choline o-acetyltransferase;PDBTitle: structures of ligand bound human choline acetyltransferase2 provide insight into regulation of acetylcholine synthesis
20 c3s6n2_
12.3
31
PDB header: splicingChain: 2: PDB Molecule: survival of motor neuron protein-interacting protein 1;PDB Fragment: gemin2-binding domain;
PDBTitle: crystal structure of the gemin2-binding domain of smn, gemin2 in2 complex with smd1/d2/f/e/g from human
21 c3nd5D_
not modelled
12.1
11
PDB header: transferaseChain: D: PDB Molecule: phosphopantetheine adenylyltransferase;PDBTitle: crystal structure of phosphopantetheine adenylyltransferase (ppat)2 from enterococcus faecalis
22 d2oa4a1
not modelled
11.6
41
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: SPO1678-like23 c3htxA_
not modelled
11.4
22
PDB header: transferase/rnaChain: A: PDB Molecule: hen1;PDBTitle: crystal structure of small rna methyltransferase hen1
24 d1vlha_
not modelled
11.1
14
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Adenylyltransferase25 c3frwF_
not modelled
10.8
13
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative trp repressor protein;PDBTitle: crystal structure of putative trpr protein from ruminococcus obeum
26 d1tfua_
not modelled
10.3
14
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Adenylyltransferase27 d1od6a_
not modelled
9.7
7
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Adenylyltransferase28 c3h0gP_
not modelled
9.7
14
PDB header: transcriptionChain: P: PDB Molecule: dna-directed rna polymerase ii subunit rpb4;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
29 c2rbgB_
not modelled
9.5
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative uncharacterized protein st0493;PDBTitle: crystal structure of hypothetical protein(st0493) from2 sulfolobus tokodaii
30 d1t07a_
not modelled
9.3
14
Fold: YggX-likeSuperfamily: YggX-likeFamily: YggX-like31 d1ckma1
not modelled
9.3
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA ligase/mRNA capping enzyme postcatalytic domain32 c2l3lA_
not modelled
8.7
20
PDB header: chaperoneChain: A: PDB Molecule: tubulin-specific chaperone c;PDBTitle: the solution structure of the n-terminal domain of human tubulin2 binding cofactor c reveals a platform for the interaction with ab-3 tubulin
33 d1qjca_
not modelled
8.6
11
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Adenylyltransferase34 c2l5aA_
not modelled
8.2
40
PDB header: nuclear proteinChain: A: PDB Molecule: histone h3-like centromeric protein cse4, protein scm3,PDBTitle: structural basis for recognition of centromere specific histone h32 variant by nonhistone scm3
35 c3f3mA_
not modelled
8.1
14
PDB header: transferaseChain: A: PDB Molecule: phosphopantetheine adenylyltransferase;PDBTitle: six crystal structures of two phosphopantetheine2 adenylyltransferases reveal an alternative ligand binding3 mode and an associated structural change
36 d1kq8a_
not modelled
8.0
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Forkhead DNA-binding domain37 c1kq8A_
not modelled
8.0
20
PDB header: transcriptionChain: A: PDB Molecule: hepatocyte nuclear factor 3 forkhead homolog 1;PDBTitle: solution structure of winged helix protein hfh-1
38 d1kama_
not modelled
7.5
21
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Adenylyltransferase39 d1o6ba_
not modelled
7.5
11
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Adenylyltransferase40 c2gutA_
not modelled
7.5
33
PDB header: transcriptionChain: A: PDB Molecule: arc/mediator, positive cofactor 2 glutamine/q-PDBTitle: solution structure of the trans-activation domain of the2 human co-activator arc105
41 c3kevA_
not modelled
7.4
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: galieria sulfuraria dcun1 domain-containing protein;PDBTitle: x-ray crystal structure of a dcun1 domain-containing protein from2 galdieria sulfuraria
42 c3ocfB_
not modelled
7.1
19
PDB header: lyaseChain: B: PDB Molecule: fumarate lyase:delta crystallin;PDBTitle: crystal structure of fumarate lyase:delta crystallin from brucella2 melitensis in native form
43 d1xs8a_
not modelled
7.0
21
Fold: YggX-likeSuperfamily: YggX-likeFamily: YggX-like44 c3krbB_
not modelled
6.4
17
PDB header: oxidoreductaseChain: B: PDB Molecule: aldose reductase;PDBTitle: structure of aldose reductase from giardia lamblia at 1.75a resolution
45 d1xsza1
not modelled
6.4
19
Fold: alpha-alpha superhelixSuperfamily: Sec7 domainFamily: Sec7 domain46 c2jrtA_
not modelled
6.2
41
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr solution structure of the protein coded by gene2 rhos4_12090 of rhodobacter sphaeroides. northeast3 structural genomics target rhr5
47 d1re0b_
not modelled
6.0
28
Fold: alpha-alpha superhelixSuperfamily: Sec7 domainFamily: Sec7 domain48 c1plpA_
not modelled
6.0
47
PDB header: membrane proteinChain: A: PDB Molecule: phospholamban;PDBTitle: solution structure of the cytoplasmic domain of2 phospholamban
49 d1ku1a_
not modelled
5.8
24
Fold: alpha-alpha superhelixSuperfamily: Sec7 domainFamily: Sec7 domain50 c3lyvF_
not modelled
5.8
13
PDB header: chaperoneChain: F: PDB Molecule: ribosome-associated factor y;PDBTitle: crystal structure of a domain of ribosome-associated factor y from2 streptococcus pyogenes serotype m6. northeast structural genomics3 consortium target id dr64a
51 d1wpga4
not modelled
5.7
17
Fold: Calcium ATPase, transmembrane domain MSuperfamily: Calcium ATPase, transmembrane domain MFamily: Calcium ATPase, transmembrane domain M52 c2jvnA_
not modelled
5.6
23
PDB header: transferaseChain: A: PDB Molecule: poly [adp-ribose] polymerase 1;PDBTitle: domain c of human parp-1
53 d1ppjj_
not modelled
5.5
26
Fold: Single transmembrane helixSuperfamily: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)54 d1rj9a1
not modelled
5.4
26
Fold: Four-helical up-and-down bundleSuperfamily: Domain of the SRP/SRP receptor G-proteinsFamily: Domain of the SRP/SRP receptor G-proteins55 c1wwuA_
not modelled
5.4
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein flj21935;PDBTitle: solution structure of the sam_pnt domain of human protein2 flj21935
56 d2r09a1
not modelled
5.4
19
Fold: alpha-alpha superhelixSuperfamily: Sec7 domainFamily: Sec7 domain57 d1ud0a_
not modelled
5.3
18
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Heat shock protein 70kD (HSP70), C-terminal subdomainFamily: Heat shock protein 70kD (HSP70), C-terminal subdomain58 d1r8se_
not modelled
5.3
7
Fold: alpha-alpha superhelixSuperfamily: Sec7 domainFamily: Sec7 domain59 c3d9wA_
not modelled
5.1
25
PDB header: transferaseChain: A: PDB Molecule: putative acetyltransferase;PDBTitle: crystal structure analysis of nocardia farcinica arylamine2 n-acetyltransferase
60 d1ppjw_
not modelled
5.1
26
Fold: Single transmembrane helixSuperfamily: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)61 d1jyoe_
not modelled
5.1
20
Fold: Non-globular alpha+beta subunits of globular proteinsSuperfamily: Non-globular alpha+beta subunits of globular proteinsFamily: Virulence effector SptP domain62 d1uhra_
not modelled
5.0
27
Fold: SWIB/MDM2 domainSuperfamily: SWIB/MDM2 domainFamily: SWIB/MDM2 domain