| 1 |
|
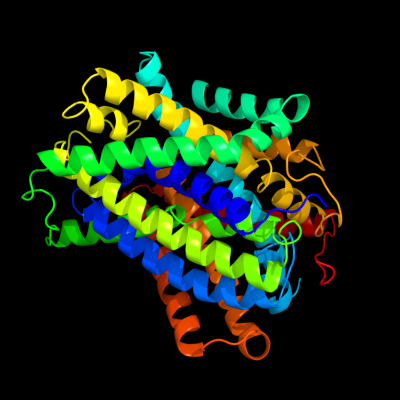
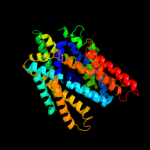
PDB 3gia chain A
Region: 12 - 455
Aligned: 421
Modelled: 421
Confidence: 100.0%
Identity: 16%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
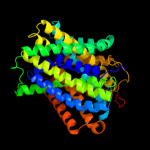
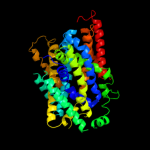
PDB 3lrc chain C
Region: 12 - 454
Aligned: 404
Modelled: 404
Confidence: 100.0%
Identity: 17%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
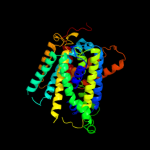
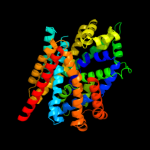
PDB 2jln chain A
Region: 3 - 457
Aligned: 442
Modelled: 442
Confidence: 100.0%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
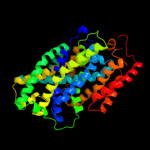
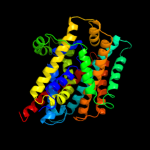
PDB 2xq2 chain A
Region: 13 - 456
Aligned: 428
Modelled: 428
Confidence: 99.6%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 5 |
|
PDB 3dh4 chain A
Region: 13 - 455
Aligned: 428
Modelled: 428
Confidence: 99.5%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 16 - 453
Aligned: 430
Modelled: 430
Confidence: 98.0%
Identity: 14%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 12 - 391
Aligned: 369
Modelled: 371
Confidence: 97.6%
Identity: 9%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 87 - 391
Aligned: 293
Modelled: 305
Confidence: 89.9%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 2rdd chain B
Region: 432 - 457
Aligned: 26
Modelled: 26
Confidence: 12.0%
Identity: 15%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 10 |
|
PDB 3rko chain F
Region: 331 - 456
Aligned: 126
Modelled: 126
Confidence: 7.2%
Identity: 11%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2