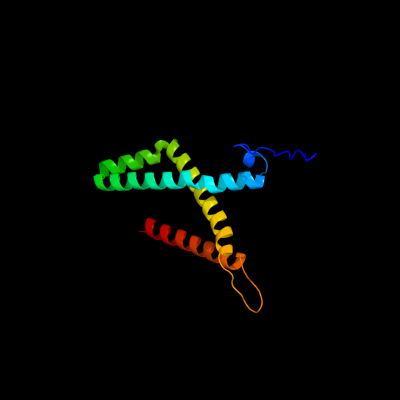
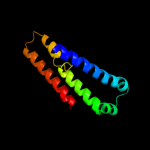
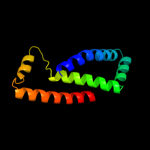
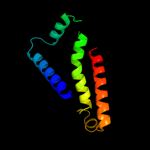
1 d1kf6c_
100.0
100
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)2 d1nekd_
96.8
20
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)3 d1nekc_
89.7
15
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)4 d1kf6d_
75.1
17
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)5 c1yq3C_
68.7
13
PDB header: oxidoreductaseChain: C: PDB Molecule: succinate dehydrogenase cytochrome b, large subunit;PDBTitle: avian respiratory complex ii with oxaloacetate and ubiquinone
6 d2bs2c1
32.7
12
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Fumarate reductase respiratory complex cytochrome b subunit, FrdC7 c2b1wA_
26.6
30
PDB header: apoptosisChain: A: PDB Molecule: caspase recruitment domain protein 4;PDBTitle: solution structure of the nod1 caspase activating and recruitment2 domain
8 d1lg7a_
23.7
36
Fold: VSV matrix proteinSuperfamily: VSV matrix proteinFamily: VSV matrix protein9 c2w2rA_
20.5
45
PDB header: viral proteinChain: A: PDB Molecule: matrix protein;PDBTitle: structure of the vesicular stomatitis virus matrix protein
10 d3ygsp_
20.0
25
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD11 c2nz7A_
19.0
33
PDB header: apoptosisChain: A: PDB Molecule: caspase recruitment domain-containing protein 4;PDBTitle: crystal structure analysis of caspase-recruitment domain2 (card) of nod1
12 c3katA_
17.5
13
PDB header: apoptosisChain: A: PDB Molecule: nacht, lrr and pyd domains-containing protein 1;PDBTitle: crystal structure of the card domain of the human nlrp12 protein, northeast structural genomics consortium target3 hr3486e
13 d3crda_
15.4
44
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD14 d1dgna_
14.1
25
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD15 c2kxwB_
13.5
44
PDB header: calcium-binding protein/metal transportChain: B: PDB Molecule: sodium channel protein type 2 subunit alpha;PDBTitle: structure of the c-domain fragment of apo calmodulin bound to the iq2 motif of nav1.2
16 d1y9ba1
12.7
20
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: VCA0319-like17 c1p58F_
11.8
16
PDB header: virusChain: F: PDB Molecule: envelope protein m;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
18 c2q0oC_
10.9
11
PDB header: transcriptionChain: C: PDB Molecule: probable transcriptional repressor tram;PDBTitle: crystal structure of an anti-activation complex in bacterial quorum2 sensing
19 c2h3oA_
9.9
24
PDB header: membrane proteinChain: A: PDB Molecule: merf;PDBTitle: structure of merft, a membrane protein with two trans-2 membrane helices
20 d1cy5a_
9.7
19
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD21 d1bw6a_
not modelled
9.5
38
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding22 c1wazA_
not modelled
8.4
24
PDB header: transport proteinChain: A: PDB Molecule: merf;PDBTitle: nmr structure determination of the bacterial mercury2 transporter, merf, in micelles
23 d2axtk1
not modelled
7.9
38
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein K, PsbKFamily: PsbK-like24 c2y69W_
not modelled
7.1
19
PDB header: electron transportChain: W: PDB Molecule: cytochrome c oxidase polypeptide 7a1;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
25 d1kcfa2
not modelled
7.0
23
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Mitochondrial resolvase ydc2 catalytic domain26 c2voyB_
not modelled
6.9
15
PDB header: hydrolaseChain: B: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
27 c1kcfB_
not modelled
6.0
23
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical 30.2 kd protein c25g10.02 inPDBTitle: crystal structure of the yeast mitochondrial holliday2 junction resolvase, ydc2
28 c2l9mA_
not modelled
5.9
28
PDB header: apoptosis inhibitorChain: A: PDB Molecule: baculoviral iap repeat-containing protein 2;PDBTitle: structure of ciap1 card
29 d1ic8a2
not modelled
5.9
30
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: POU-specific domain30 c2zidA_
not modelled
5.4
42
PDB header: hydrolaseChain: A: PDB Molecule: dextran glucosidase;PDBTitle: crystal structure of dextran glucosidase e236q complex with2 isomaltotriose