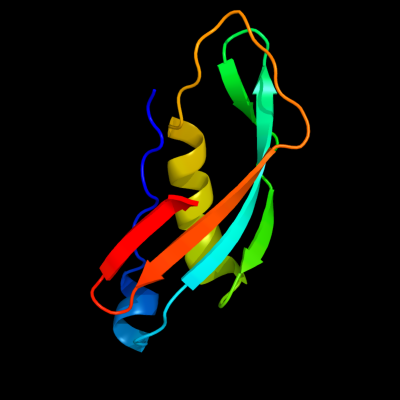
1 c1xx3A_
99.8
100
PDB header: transport proteinChain: A: PDB Molecule: tonb protein;PDBTitle: solution structure of escherichia coli tonb-ctd
2 d1u07a_
99.8
100
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB3 c2grxC_
99.8
100
PDB header: metal transportChain: C: PDB Molecule: protein tonb;PDBTitle: crystal structure of tonb in complex with fhua, e. coli2 outer membrane receptor for ferrichrome
4 d2gskb1
99.8
100
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB5 d1ihra_
99.7
100
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB6 c2k9kA_
99.7
31
PDB header: metal transportChain: A: PDB Molecule: tonb2;PDBTitle: molecular characterization of the tonb2 protein from vibrio2 anguillarum
7 d1lr0a_
99.4
22
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TolA8 c2x9aB_
99.2
11
PDB header: viral proteinChain: B: PDB Molecule: membrane spanning protein, required for outer membranePDBTitle: crystal structure of g3p from phage if1 in complex with its2 coreceptor, the c-terminal domain of tola
9 d1tola2
99.1
10
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TolA10 c1tolA_
98.7
10
PDB header: viral proteinChain: A: PDB Molecule: protein (fusion protein consisting of minor coatPDBTitle: fusion of n-terminal domain of the minor coat protein from2 gene iii in phage m13, and c-terminal domain of e. coli3 protein-tola
11 c1ybxA_
64.2
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: conserved hypothetical protein cth-383 from clostridium thermocellum
12 c3onrI_
60.8
21
PDB header: metal binding proteinChain: I: PDB Molecule: protein transport protein sece2;PDBTitle: crystal structure of the calcium chelating immunodominant antigen,2 calcium dodecin (rv0379),from mycobacterium tuberculosis with a novel3 calcium-binding site
13 c2h66G_
60.0
9
PDB header: structural genomics/oxidoreductaseChain: G: PDB Molecule: pv-pf14_0368;PDBTitle: the crystal structure of plasmodium vivax 2-cys2 peroxiredoxin
14 c3kebB_
57.6
6
PDB header: oxidoreductaseChain: B: PDB Molecule: probable thiol peroxidase;PDBTitle: thiol peroxidase from chromobacterium violaceum
15 d1j8ba_
57.6
17
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like16 d1puga_
52.2
22
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like17 d2cv4a1
50.7
11
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like18 c3drnB_
47.4
20
PDB header: oxidoreductaseChain: B: PDB Molecule: peroxiredoxin, bacterioferritin comigratory proteinPDBTitle: the crystal structure of bcp1 from sulfolobus sulfataricus
19 d2ux9a1
45.5
25
Fold: Dodecin subunit-likeSuperfamily: Dodecin-likeFamily: Dodecin-like20 d1we0a1
43.6
21
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like21 d1zyea1
not modelled
42.8
13
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like22 d2h01a1
not modelled
41.6
9
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like23 c2vxaL_
not modelled
40.3
32
PDB header: flavoproteinChain: L: PDB Molecule: dodecin;PDBTitle: h.halophila dodecin in complex with riboflavin
24 d1e2ya_
not modelled
38.3
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like25 c2bmxB_
not modelled
36.4
17
PDB header: oxidoreductaseChain: B: PDB Molecule: alkyl hydroperoxidase c;PDBTitle: mycobacterium tuberculosis ahpc
26 d1qq2a_
not modelled
33.8
11
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like27 d1qmva_
not modelled
33.5
9
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like28 c3hd7A_
not modelled
31.6
15
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
29 d1n8ja_
not modelled
31.4
13
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like30 c3oqtP_
not modelled
31.1
14
PDB header: flavoproteinChain: P: PDB Molecule: rv1498a protein;PDBTitle: crystal structure of rv1498a protein from mycobacterium tuberculosis
31 d1uula_
not modelled
30.9
9
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like32 d1m98a2
not modelled
30.5
23
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Orange carotenoid protein, C-terminal domain33 c3dtdI_
not modelled
29.6
22
PDB header: structural genomics, unknown functionChain: I: PDB Molecule: invasion-associated protein b;PDBTitle: crystal structure of invasion associated protein b from bartonella2 henselae
34 d1psqa_
not modelled
29.1
12
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like35 d1xvqa_
not modelled
29.0
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like36 c2ii1A_
not modelled
28.7
17
PDB header: hydrolaseChain: A: PDB Molecule: acetamidase;PDBTitle: crystal structure of acetamidase (10172637) from bacillus halodurans2 at 1.95 a resolution
37 c2kzxA_
not modelled
28.7
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of a3dht5 from clostridium thermocellum,2 northeast structural genomics consortium target cmr116
38 c2yzhD_
not modelled
26.1
24
PDB header: oxidoreductaseChain: D: PDB Molecule: probable thiol peroxidase;PDBTitle: crystal structure of peroxiredoxin-like protein from aquifex aeolicus
39 c2xpdC_
not modelled
25.3
6
PDB header: oxidoreductaseChain: C: PDB Molecule: thiol peroxidase;PDBTitle: reduced thiol peroxidase (tpx) from yersinia pseudotuberculosis
40 d1ijda_
not modelled
24.5
21
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits41 d2cx4a1
not modelled
23.6
24
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like42 d1nkza_
not modelled
22.1
17
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits43 d2bmxa1
not modelled
21.9
17
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like44 d2vnud4
not modelled
21.3
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: RNB domain-like45 c2c0dA_
not modelled
20.9
11
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin peroxidase 2;PDBTitle: structure of the mitochondrial 2-cys peroxiredoxin from2 plasmodium falciparum
46 d1zofa1
not modelled
20.7
11
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like47 c3o6uB_
not modelled
20.4
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein cpe2226;PDBTitle: crystal structure of cpe2226 protein from clostridium perfringens.2 northeast structural genomics consortium target cpr195
48 c2r7fA_
not modelled
18.9
27
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease ii family protein;PDBTitle: crystal structure of ribonuclease ii family protein from deinococcus2 radiodurans, hexagonal crystal form. northeast structural genomics3 target drr63
49 d2r7da2
not modelled
18.4
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: RNB domain-like50 c3lwxA_
not modelled
18.1
9
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh:ubiquinone oxidoreductase, na translocating,PDBTitle: crystal structure of na(+)-translocating nadh-quinone2 reductase subunit c (yp_001302508.1) from parabacteroides3 distasonis atcc 8503 at 1.10 a resolution
51 d1knga_
not modelled
18.0
12
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like52 c1zyeL_
not modelled
17.9
15
PDB header: oxidoreductaseChain: L: PDB Molecule: thioredoxin-dependent peroxide reductase;PDBTitle: crystal strucutre analysis of bovine mitochondrial peroxiredoxin iii
53 c2egtA_
not modelled
15.5
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein aq_1549;PDBTitle: crystal structure of hypothetical protein (aq1549) from aquifex2 aeolicus
54 c2jszA_
not modelled
14.8
15
PDB header: oxidoreductaseChain: A: PDB Molecule: probable thiol peroxidase;PDBTitle: solution structure of tpx in the reduced state
55 d1pugb_
not modelled
13.8
22
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like56 c3eytA_
not modelled
13.6
6
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein spoa0173;PDBTitle: crystal structure of thioredoxin-like superfamily protein spoa0173
57 c1h2aL_
not modelled
12.8
30
PDB header: oxidoreductaseChain: L: PDB Molecule: hydrogenase;PDBTitle: single crystals of hydrogenase from desulfovibrio vulgaris
58 d1iwla_
not modelled
12.7
10
Fold: LolA-like prokaryotic lipoproteins and lipoprotein localization factorsSuperfamily: Prokaryotic lipoproteins and lipoprotein localization factorsFamily: Outer-membrane lipoproteins carrier protein LolA59 c2ywnA_
not modelled
12.3
7
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxiredoxin-like protein;PDBTitle: crystal structure of peroxiredoxin-like protein from2 sulfolobus tokodaii
60 c1zofB_
not modelled
11.1
12
PDB header: oxidoreductaseChain: B: PDB Molecule: alkyl hydroperoxide-reductase;PDBTitle: crystal structure of alkyl hydroperoxide-reductase (ahpc)2 from helicobacter pylori
61 d2idla1
not modelled
11.0
14
Fold: eIF1-likeSuperfamily: TM1457-likeFamily: TM1457-like62 d1xvwa1
not modelled
10.9
14
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like63 d2zcta1
not modelled
10.7
12
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like64 d1wuil1
not modelled
10.5
31
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit65 c3mjjD_
not modelled
10.3
19
PDB header: hydrolaseChain: D: PDB Molecule: predicted acetamidase/formamidase;PDBTitle: crystal structure analysis of a recombinant predicted2 acetamidase/formamidase from the thermophile thermoanaerobacter3 tengcongensis
66 c2b7kD_
not modelled
10.1
6
PDB header: metal binding proteinChain: D: PDB Molecule: sco1 protein;PDBTitle: crystal structure of yeast sco1
67 d2ix0a4
not modelled
9.6
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: RNB domain-like68 d1xcca_
not modelled
9.5
10
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like69 c3myrB_
not modelled
9.2
18
PDB header: oxidoreductaseChain: B: PDB Molecule: nickel-dependent hydrogenase large subunit;PDBTitle: crystal structure of [nife] hydrogenase from allochromatium vinosum in2 its ni-a state
70 c2fug4_
not modelled
8.9
8
PDB header: oxidoreductaseChain: 4: PDB Molecule: nadh-quinone oxidoreductase chain 4;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
71 d2fug41
not modelled
8.9
8
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nqo4-like72 c3gknA_
not modelled
8.6
26
PDB header: oxidoreductaseChain: A: PDB Molecule: bacterioferritin comigratory protein;PDBTitle: insights into the alkyl peroxide reduction activity of xanthomonas2 campestris bacterioferritin comigratory protein from the trapped3 intermediate/ligand complex structures
73 c1vqzA_
not modelled
8.6
21
PDB header: ligaseChain: A: PDB Molecule: lipoate-protein ligase, putative;PDBTitle: crystal structure of a putative lipoate-protein ligase a (sp_1160)2 from streptococcus pneumoniae tigr4 at 1.99 a resolution
74 c2vnuD_
not modelled
8.2
14
PDB header: hydrolase/rnaChain: D: PDB Molecule: exosome complex exonuclease rrp44;PDBTitle: crystal structure of sc rrp44
75 c2kl8A_
not modelled
8.1
22
PDB header: de novo proteinChain: A: PDB Molecule: or15;PDBTitle: solution nmr structure of de novo designed ferredoxin-like2 fold protein, northeast structural genomics consortium3 target or15
76 c3kh7A_
not modelled
7.9
6
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbe;PDBTitle: crystal structure of the periplasmic soluble domain of reduced ccmg2 from pseudomonas aeruginosa
77 c3razA_
not modelled
7.8
15
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin-related protein;PDBTitle: the crystal structure of thioredoxin-related protein from neisseria2 meningitidis serogroup b
78 d2b7ka1
not modelled
7.7
6
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like79 d1yexa1
not modelled
7.6
15
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like80 c2kncA_
not modelled
7.4
19
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
81 d1qxha_
not modelled
7.4
12
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like82 d1xrda1
not modelled
7.2
16
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits83 c2nr1A_
not modelled
7.2
39
PDB header: receptorChain: A: PDB Molecule: nr1 m2;PDBTitle: transmembrane segment 2 of nmda receptor nr1, nmr, 102 structures
84 d1rjlc_
not modelled
7.1
31
Fold: open-sided beta-meanderSuperfamily: Outer surface proteinFamily: Outer surface protein85 d1yq9h1
not modelled
6.9
30
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit86 c2v1mA_
not modelled
6.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: glutathione peroxidase;PDBTitle: crystal structure of schistosoma mansoni glutathione2 peroxidase
87 d1q98a_
not modelled
6.1
11
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like88 d1frfl_
not modelled
6.1
30
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit89 c3f8xD_
not modelled
6.0
7
PDB header: isomeraseChain: D: PDB Molecule: putative delta-5-3-ketosteroid isomerase;PDBTitle: crystal structure of a putative delta-5-3-ketosteroid isomerase2 (eca2236) from pectobacterium atrosepticum scri1043 at 1.55 a3 resolution
90 d1e3db_
not modelled
5.8
25
Fold: HydB/Nqo4-likeSuperfamily: HydB/Nqo4-likeFamily: Nickel-iron hydrogenase, large subunit91 c2boyC_
not modelled
5.7
12
PDB header: oxidoreductaseChain: C: PDB Molecule: 3-chlorocatechol 1,2-dioxygenase;PDBTitle: crystal structure of 3-chlorocatechol 1,2-dioxygenase from2 rhodococcus opacus 1cp
92 c3fljA_
not modelled
5.3
3
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein conserved in bacteria with aPDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 with a cystatin-like fold (yp_168589.1) from silicibacter pomeroyi3 dss-3 at 2.00 a resolution
93 c2qtsA_
not modelled
5.1
8
PDB header: membrane proteinChain: A: PDB Molecule: acid-sensing ion channel;PDBTitle: structure of an acid-sensing ion channel 1 at 1.9 a resolution and low2 ph
94 c2wpnB_
not modelled
5.0
33
PDB header: oxidoreductaseChain: B: PDB Molecule: periplasmic [nifese] hydrogenase, large subunit,PDBTitle: structure of the oxidised, as-isolated nifese hydrogenase2 from d. vulgaris hildenborough
95 c2rliA_
not modelled
5.0
19
PDB header: metal transportChain: A: PDB Molecule: sco2 protein homolog, mitochondrial;PDBTitle: solution structure of cu(i) human sco2
96 d3bzka5
not modelled
5.0
5
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like