1 c2j6pF_
99.9
21
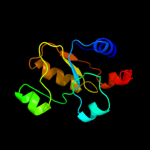
PDB header: oxidoreductaseChain: F: PDB Molecule: sb(v)-as(v) reductase;PDBTitle: structure of as-sb reductase from leishmania major
2 c3i2vA_
99.8
21
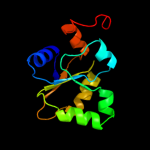
PDB header: transferaseChain: A: PDB Molecule: adenylyltransferase and sulfurtransferase mocs3;PDBTitle: crystal structure of human mocs3 rhodanese-like domain
3 c3tp9B_
99.8
26
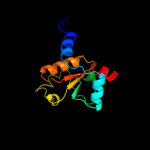
PDB header: hydrolaseChain: B: PDB Molecule: beta-lactamase and rhodanese domain protein;PDBTitle: crystal structure of alicyclobacillus acidocaldarius protein with2 beta-lactamase and rhodanese domains
4 d1c25a_
99.8
22
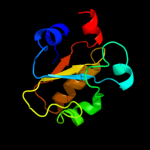
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Cell cycle control phosphatase, catalytic domain5 c3d1pA_
99.8
17
PDB header: transferaseChain: A: PDB Molecule: putative thiosulfate sulfurtransferase yor285w;PDBTitle: atomic resolution structure of uncharacterized protein from2 saccharomyces cerevisiae
6 c3f4aA_
99.8
15
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein ygr203w;PDBTitle: structure of ygr203w, a yeast protein tyrosine phosphatase2 of the rhodanese family
7 c2hhgA_
99.8
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein rpa3614;PDBTitle: structure of protein of unknown function rpa3614, possible tyrosine2 phosphatase, from rhodopseudomonas palustris cga009
8 d1yt8a4
99.8
22
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)9 d1yt8a1
99.8
21
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)10 c3ilmD_
99.8
23
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: alr3790 protein;PDBTitle: crystal structure of the alr3790 protein from anabaena sp. northeast2 structural genomics consortium target nsr437h
11 c3fojA_
99.8
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of ssp1007 from staphylococcus2 saprophyticus subsp. saprophyticus. northeast structural3 genomics target syr101a.
12 d1gmxa_
99.8
20
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Single-domain sulfurtransferase13 c3icrA_
99.8
26
PDB header: oxidoreductaseChain: A: PDB Molecule: coenzyme a-disulfide reductase;PDBTitle: crystal structure of oxidized bacillus anthracis coadr-rhd
14 d1ymka1
99.8
20
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Cell cycle control phosphatase, catalytic domain15 c1yt8A_
99.8
14
PDB header: transferaseChain: A: PDB Molecule: thiosulfate sulfurtransferase;PDBTitle: crystal structure of thiosulfate sulfurtransferase from pseudomonas2 aeruginosa
16 c2dcqA_
99.8
15
PDB header: unknown functionChain: A: PDB Molecule: putative protein at4g01050;PDBTitle: fully automated nmr structure determination of the2 rhodanese homology domain at4g01050(175-295) from3 arabidopsis thaliana
17 c2fsxA_
99.8
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cog0607: rhodanese-related sulfurtransferase;PDBTitle: crystal structure of rv0390 from m. tuberculosis
18 d1yt8a2
99.8
18
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)19 d1qxna_
99.8
19
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Single-domain sulfurtransferase20 c1e0cA_
99.8
20
PDB header: sulfurtransferaseChain: A: PDB Molecule: sulfurtransferase;PDBTitle: sulfurtransferase from azotobacter vinelandii
21 c3emeA_
not modelled
99.7
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: rhodanese-like domain protein;PDBTitle: crystal structure of rhodanese-like domain protein from2 staphylococcus aureus
22 c3aaxB_
not modelled
99.7
19
PDB header: transferaseChain: B: PDB Molecule: putative thiosulfate sulfurtransferase;PDBTitle: crystal structure of probable thiosulfate sulfurtransferase2 cysa3 (rv3117) from mycobacterium tuberculosis: monoclinic3 form
23 c3gk5A_
not modelled
99.7
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized rhodanese-related proteinPDBTitle: crystal structure of rhodanese-related protein (tvg0868615)2 from thermoplasma volcanium, northeast structural genomics3 consortium target tvr109a
24 c1urhA_
not modelled
99.7
27
PDB header: transferaseChain: A: PDB Molecule: 3-mercaptopyruvate sulfurtransferase;PDBTitle: the "rhodanese" fold and catalytic mechanism of2 3-mercaptopyruvate sulfotransferases: crystal structure3 of ssea from escherichia coli
25 d1yt8a3
not modelled
99.7
15
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)26 c3op3A_
not modelled
99.7
23
PDB header: hydrolaseChain: A: PDB Molecule: m-phase inducer phosphatase 3;PDBTitle: crystal structure of cell division cycle 25c protein isoform a from2 homo sapiens
27 d1tq1a_
not modelled
99.7
22
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Single-domain sulfurtransferase28 c3hzuA_
not modelled
99.7
20
PDB header: transferaseChain: A: PDB Molecule: thiosulfate sulfurtransferase ssea;PDBTitle: crystal structure of probable thiosulfate sulfurtransferase ssea2 (rhodanese) from mycobacterium tuberculosis
29 c3g5jA_
not modelled
99.7
27
PDB header: nucleotide binding proteinChain: A: PDB Molecule: putative atp/gtp binding protein;PDBTitle: crystal structure of n-terminal domain of putative atp/gtp binding2 protein from clostridium difficile 630
30 c2uzqE_
not modelled
99.7
21
PDB header: hydrolaseChain: E: PDB Molecule: m-phase inducer phosphatase 2;PDBTitle: protein phosphatase, new crystal form
31 c3k9rA_
not modelled
99.7
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: alr3790 protein;PDBTitle: x-ray structure of the rhodanese-like domain of the alr3790 protein2 from anabaena sp. northeast structural genomics consortium target3 nsr437c.
32 c3ntaA_
not modelled
99.7
28
PDB header: oxidoreductaseChain: A: PDB Molecule: fad-dependent pyridine nucleotide-disulphidePDBTitle: structure of the shewanella loihica pv-4 nadh-dependent persulfide2 reductase
33 d1e0ca1
not modelled
99.7
20
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)34 d1uara2
not modelled
99.7
28
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)35 c1uarA_
not modelled
99.7
24
PDB header: transferaseChain: A: PDB Molecule: rhodanese;PDBTitle: crystal structure of rhodanese from thermus thermophilus hb8
36 c1boiA_
not modelled
99.7
16
PDB header: transferaseChain: A: PDB Molecule: rhodanese;PDBTitle: n-terminally truncated rhodanese
37 c3ippA_
not modelled
99.7
20
PDB header: transferaseChain: A: PDB Molecule: putative thiosulfate sulfurtransferase ynje;PDBTitle: crystal structure of sulfur-free ynje
38 d1t3ka_
not modelled
99.7
16
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Cell cycle control phosphatase, catalytic domain39 c3olhA_
not modelled
99.7
20
PDB header: transferaseChain: A: PDB Molecule: 3-mercaptopyruvate sulfurtransferase;PDBTitle: human 3-mercaptopyruvate sulfurtransferase
40 c2jtqA_
not modelled
99.7
25
PDB header: transferaseChain: A: PDB Molecule: phage shock protein e;PDBTitle: rhodanese from e.coli
41 d1rhsa1
not modelled
99.7
15
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)42 c1wv9B_
not modelled
99.6
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: rhodanese homolog tt1651;PDBTitle: crystal structure of rhodanese homolog tt1651 from an2 extremely thermophilic bacterium thermus thermophilus hb8
43 c3nhvE_
not modelled
99.6
26
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: bh2092 protein;PDBTitle: crystal structure of bh2092 protein from bacillus halodurans,2 northeast structural genomics consortium target bhr228f
44 c2k0zA_
not modelled
99.6
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein hp1203;PDBTitle: solution nmr structure of protein hp1203 from helicobacter pylori2 26695. northeast structural genomics consortium (nesg) target3 pt1/ontario center for structural proteomics target hp1203
45 d1urha1
not modelled
99.6
24
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)46 d1e0ca2
not modelled
99.6
24
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)47 d1okga2
not modelled
99.6
21
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)48 d1rhsa2
not modelled
99.6
20
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)49 d1uara1
not modelled
99.6
24
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)50 c2vswB_
not modelled
99.6
22
PDB header: hydrolaseChain: B: PDB Molecule: dual specificity protein phosphatase 16;PDBTitle: the structure of the rhodanese domain of the human dual2 specificity phosphatase 16
51 c3r2uC_
not modelled
99.4
24
PDB header: hydrolaseChain: C: PDB Molecule: metallo-beta-lactamase family protein;PDBTitle: 2.1 angstrom resolution crystal structure of metallo-beta-lactamase2 from staphylococcus aureus subsp. aureus col
52 d1urha2
not modelled
99.4
20
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)53 d1okga1
not modelled
99.4
19
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)54 c2oucB_
not modelled
99.4
16
PDB header: hydrolaseChain: B: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of the map kinase binding domain of mkp5
55 d2gwfa1
not modelled
99.4
16
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Ubiquitin carboxyl-terminal hydrolase 8, USP856 c2eg4B_
not modelled
99.4
18
PDB header: transferaseChain: B: PDB Molecule: probable thiosulfate sulfurtransferase;PDBTitle: crystal structure of probable thiosulfate sulfurtransferase
57 d1whba_
not modelled
99.4
15
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Ubiquitin carboxyl-terminal hydrolase 8, USP858 c1okgA_
not modelled
99.3
20
PDB header: transferaseChain: A: PDB Molecule: possible 3-mercaptopyruvate sulfurtransferase;PDBTitle: 3-mercaptopyruvate sulfurtransferase from leishmania major
59 d1hzma_
not modelled
99.3
19
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Cell cycle control phosphatase, catalytic domain60 d1urra_
95.8
15
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like61 c3flhC_
not modelled
95.7
24
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein lp_1913;PDBTitle: crystal structure of lp_1913 protein from lactobacillus2 plantarum,northeast structural genomics consortium target3 lpr140b
62 c2gv1A_
95.6
14
PDB header: hydrolaseChain: A: PDB Molecule: probable acylphosphatase;PDBTitle: nmr solution structure of the acylphosphatase from2 eschaerichia coli
63 d1d5ra2
not modelled
95.5
10
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like64 d2acya_
95.5
15
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like65 c3br8A_
94.9
10
PDB header: hydrolaseChain: A: PDB Molecule: probable acylphosphatase;PDBTitle: crystal structure of acylphosphatase from bacillus subtilis
66 c3rgqA_
not modelled
94.6
15
PDB header: hydrolaseChain: A: PDB Molecule: protein-tyrosine phosphatase mitochondrial 1;PDBTitle: crystal structure of ptpmt1 in complex with pi(5)p
67 d1apsa_
94.4
12
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like68 d1ulra_
94.4
14
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like69 c3gxgA_
not modelled
94.3
13
PDB header: hydrolaseChain: A: PDB Molecule: putative phosphatase (duf442);PDBTitle: crystal structure of putative phosphatase (duf442) (yp_001181608.1)2 from shewanella putrefaciens cn-32 at 1.60 a resolution
70 d1w2ia_
not modelled
94.1
18
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like71 c2f46A_
not modelled
94.1
17
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a putative phosphatase (nma1982) from neisseria2 meningitidis z2491 at 1.41 a resolution
72 c2bjeA_
not modelled
94.0
11
PDB header: hydrolaseChain: A: PDB Molecule: acylphosphatase;PDBTitle: acylphosphatase from sulfolobus solfataricus. monclinic p212 space group
73 c3awfC_
not modelled
93.8
18
PDB header: hydrolase, membrane proteinChain: C: PDB Molecule: voltage-sensor containing phosphatase;PDBTitle: crystal structure of pten-like domain of ci-vsp (236-576)
74 c2imgA_
not modelled
93.5
14
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 23;PDBTitle: crystal structure of dual specificity protein phosphatase2 23 from homo sapiens in complex with ligand malate ion
75 d1xria_
not modelled
92.9
11
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like76 c1d5rA_
not modelled
92.0
10
PDB header: hydrolaseChain: A: PDB Molecule: phosphoinositide phosphotase pten;PDBTitle: crystal structure of the pten tumor suppressor
77 c1yn9B_
not modelled
88.6
17
PDB header: hydrolaseChain: B: PDB Molecule: polynucleotide 5'-phosphatase;PDBTitle: crystal structure of baculovirus rna 5'-phosphatase2 complexed with phosphate
78 d1ywfa1
not modelled
87.7
16
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Mycobacterial PtpB-like79 c3rz2B_
not modelled
87.5
13
PDB header: hydrolaseChain: B: PDB Molecule: protein tyrosine phosphatase type iva 1;PDBTitle: crystal of prl-1 complexed with peptide
80 c1fpzF_
not modelled
87.1
13
PDB header: hydrolaseChain: F: PDB Molecule: cyclin-dependent kinase inhibitor 3;PDBTitle: crystal structure analysis of kinase associated phosphatase2 (kap) with a substitution of the catalytic site cysteine3 (cys140) to a serine
81 d1rxda_
not modelled
86.4
11
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like82 d1jl3a_
not modelled
82.4
29
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases83 d1oywa3
not modelled
81.8
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain84 d1fpza_
not modelled
80.7
13
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like85 c3n0aA_
not modelled
80.1
13
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine-protein phosphatase auxilin;PDBTitle: crystal structure of auxilin (40-400)
86 c3t38B_
not modelled
79.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: arsenate reductase;PDBTitle: corynebacterium glutamicum thioredoxin-dependent arsenate reductase2 cg_arsc1'
87 c2c46B_
not modelled
78.2
13
PDB header: transferaseChain: B: PDB Molecule: mrna capping enzyme;PDBTitle: crystal structure of the human rna guanylyltransferase and2 5'-phosphatase
88 c3i32A_
77.4
7
PDB header: rna binding protein,hydrolaseChain: A: PDB Molecule: heat resistant rna dependent atpase;PDBTitle: dimeric structure of a hera helicase fragment including the c-terminal2 reca domain, the dimerization domain, and the rna binding domain
89 c2l18A_
not modelled
77.0
26
PDB header: oxidoreductaseChain: A: PDB Molecule: arsenate reductase;PDBTitle: an arsenate reductase in the phosphate binding state
90 c2vxoB_
not modelled
75.1
14
PDB header: ligaseChain: B: PDB Molecule: gmp synthase [glutamine-hydrolyzing];PDBTitle: human gmp synthetase in complex with xmp
91 d1fuka_
not modelled
75.0
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain92 d1a1va2
not modelled
74.1
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RNA helicase93 d1y4oa1
not modelled
73.8
18
Fold: Profilin-likeSuperfamily: Roadblock/LC7 domainFamily: Roadblock/LC7 domain94 c1oheA_
not modelled
73.1
17
PDB header: hydrolaseChain: A: PDB Molecule: cdc14b2 phosphatase;PDBTitle: structure of cdc14b phosphatase with a peptide ligand
95 d1jf8a_
not modelled
72.7
26
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases96 c2i6oA_
not modelled
72.1
16
PDB header: hydrolaseChain: A: PDB Molecule: sulfolobus solfataricus protein tyrosinePDBTitle: crystal structure of the complex of the archaeal sulfolobus2 ptp-fold phosphatase with phosphopeptides n-g-(p)y-k-n
97 d1yrxa1
not modelled
64.9
11
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: BLUF domain98 c3rofA_
not modelled
63.7
18
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-phosphatase ptpa;PDBTitle: crystal structure of the s. aureus protein tyrosine phosphatase ptpa
99 c1dvpA_
63.3
17
PDB header: transferaseChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosinePDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
100 d1ni5a1
not modelled
63.2
14
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: PP-loop ATPase101 c1yz4A_
not modelled
62.5
10
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase-like 15 isoform a;PDBTitle: crystal structure of dusp15
102 c2hcmA_
not modelled
62.3
13
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase;PDBTitle: crystal structure of mouse putative dual specificity phosphatase2 complexed with zinc tungstate, new york structural genomics3 consortium
103 c1k97A_
not modelled
62.2
18
PDB header: ligaseChain: A: PDB Molecule: argininosuccinate synthase;PDBTitle: crystal structure of e. coli argininosuccinate synthetase in complex2 with aspartate and citrulline
104 c3ptwA_
not modelled
61.1
14
PDB header: transferaseChain: A: PDB Molecule: malonyl coa-acyl carrier protein transacylase;PDBTitle: crystal structure of malonyl coa-acyl carrier protein transacylase2 from clostridium perfringens atcc 13124
105 c2f55C_
not modelled
60.2
17
PDB header: hydrolase/dnaChain: C: PDB Molecule: polyprotein;PDBTitle: two hepatitis c virus ns3 helicase domains complexed with2 the same strand of dna
106 c1oywA_
not modelled
59.9
19
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent dna helicase;PDBTitle: structure of the recq catalytic core
107 c2jgnB_
not modelled
58.2
12
PDB header: hydrolaseChain: B: PDB Molecule: atp-dependent rna helicase ddx3x;PDBTitle: ddx3 helicase domain
108 d1gxua_
not modelled
58.2
22
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like109 c2v1xB_
not modelled
58.1
11
PDB header: hydrolaseChain: B: PDB Molecule: atp-dependent dna helicase q1;PDBTitle: crystal structure of human recq-like dna helicase
110 d1m3ga_
not modelled
58.0
10
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like111 c1wrmA_
not modelled
58.0
11
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity phosphatase 22;PDBTitle: crystal structure of jsp-1
112 d1v3aa_
not modelled
57.9
13
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Dual specificity phosphatase-like113 d2g2ja1
not modelled
57.6
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain114 c2fekA_
not modelled
57.6
18
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-PDBTitle: structure of a protein tyrosine phosphatase
115 c2gi4A_
not modelled
57.5
39
PDB header: hydrolaseChain: A: PDB Molecule: possible phosphotyrosine protein phosphatase;PDBTitle: solution structure of the low molecular weight protein2 tyrosine phosphatase from campylobacter jejuni.
116 c2wmyH_
not modelled
56.6
21
PDB header: hydrolaseChain: H: PDB Molecule: putative acid phosphatase wzb;PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
117 c2oz5A_
not modelled
55.9
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phosphotyrosine protein phosphatase ptpb;PDBTitle: crystal structure of mycobacterium tuberculosis protein tyrosine2 phosphatase ptpb in complex with the specific inhibitor omts
118 d2j0sa2
not modelled
54.3
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain119 d1y1la_
not modelled
54.2
22
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases120 c2cwdA_
not modelled
53.2
15
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight phosphotyrosine protein phosphatase;PDBTitle: crystal structure of tt1001 protein from thermus thermophilus hb8