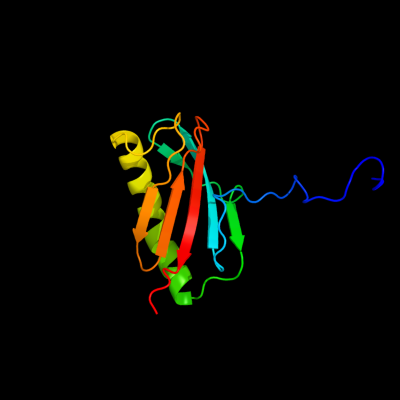
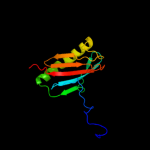
| 1 | d2inwa1
|
|
 |
100.0 |
91 |
Fold:Profilin-like
Superfamily:YeeU-like
Family:YagB/YeeU/YfjZ-like |
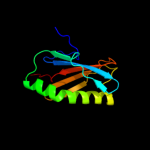
| 2 | d2h28a1
|
|
 |
100.0 |
97 |
Fold:Profilin-like
Superfamily:YeeU-like
Family:YagB/YeeU/YfjZ-like |
| 3 | d2ea9a1
|
|
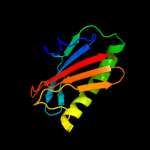 |
100.0 |
65 |
Fold:Profilin-like
Superfamily:YeeU-like
Family:YagB/YeeU/YfjZ-like |
| 4 | d1ryba_
|
|
 |
52.1 |
20 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Peptidyl-tRNA hydrolase-like
Family:Peptidyl-tRNA hydrolase-like |
| 5 | c3h3yF_
|
|
 |
46.2 |
22 |
PDB header:viral protein
Chain: F: PDB Molecule:baseplate structural protein gp6;
PDBTitle: fitting of the gp6 crystal structure into 3d cryo-em2 reconstruction of bacteriophage t4 star-shaped baseplate
|
| 6 | c2z2jA_
|
|
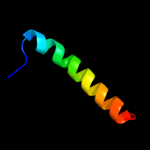 |
44.6 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from mycobacterium2 tuberculosis
|
| 7 | c3v2iA_
|
|
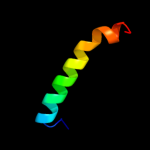 |
25.1 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: structure of a peptidyl-trna hydrolase (pth) from burkholderia2 thailandensis
|
| 8 | d2e7ga1
|
|
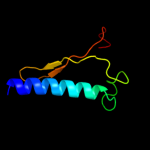 |
24.6 |
21 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Ribosome-binding factor A, RbfA
Family:Ribosome-binding factor A, RbfA |
| 9 | d2ptha_
|
|
 |
24.4 |
21 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Peptidyl-tRNA hydrolase-like
Family:Peptidyl-tRNA hydrolase-like |
| 10 | c3neaA_
|
|
 |
23.6 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from francisella2 tularensis
|
| 11 | c2yqkA_
|
|
 |
20.9 |
9 |
PDB header:transcription/apoptosis
Chain: A: PDB Molecule:arginine-glutamic acid dipeptide repeats protein;
PDBTitle: solution structure of the sant domain in arginine-glutamic2 acid dipeptide (re) repeats
|
| 12 | d1du0a_
|
|
 |
19.2 |
29 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 13 | d2qdya1
|
|
 |
17.7 |
21 |
Fold:Nitrile hydratase alpha chain
Superfamily:Nitrile hydratase alpha chain
Family:Nitrile hydratase alpha chain |
| 14 | d1lpba2
|
|
 |
16.7 |
71 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Colipase-like
Family:Colipase-like |
| 15 | d1xv2a_
|
|
 |
15.7 |
23 |
Fold:AF0104/ALDC/Ptd012-like
Superfamily:AF0104/ALDC/Ptd012-like
Family:Alpha-acetolactate decarboxylase-like |
| 16 | d1p7jb_
|
|
 |
14.5 |
29 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 17 | d1mh3a1
|
|
 |
14.4 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 18 | d1s7ea1
|
|
 |
14.3 |
31 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 19 | d1p91a_
|
|
 |
13.5 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:rRNA methyltransferase RlmA |
| 20 | d1le8a_
|
|
 |
13.0 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 21 | c3aqoD_ |
|
not modelled |
12.7 |
48 |
PDB header:membrane protein
Chain: D: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: structure and function of a membrane component secdf that enhances2 protein export
|
| 22 | c3hruA_ |
|
not modelled |
12.5 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:metalloregulator scar;
PDBTitle: crystal structure of scar with bound zn2+
|
| 23 | d2obba1 |
|
not modelled |
12.1 |
26 |
Fold:HAD-like
Superfamily:HAD-like
Family:BT0820-like |
| 24 | d1e3oc1 |
|
not modelled |
11.3 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 25 | c3a03A_ |
|
not modelled |
10.6 |
31 |
PDB header:gene regulation
Chain: A: PDB Molecule:t-cell leukemia homeobox protein 2;
PDBTitle: crystal structure of hox11l1 homeodomain
|
| 26 | c1ze2B_ |
|
not modelled |
10.5 |
16 |
PDB header:lyase/rna
Chain: B: PDB Molecule:trna pseudouridine synthase b;
PDBTitle: conformational change of pseudouridine 55 synthase upon its2 association with rna substrate
|
| 27 | c1n8sC_ |
|
not modelled |
10.2 |
43 |
PDB header:hydrolase
Chain: C: PDB Molecule:colipase ii;
PDBTitle: structure of the pancreatic lipase-colipase complex
|
| 28 | d1au7a1 |
|
not modelled |
10.0 |
31 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 29 | c2lcgA_ |
|
not modelled |
9.9 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of protein rmet_5065 from ralstonia2 metallidurans, northeast structural genomics consortium target crr115
|
| 30 | d1g3wa2 |
|
not modelled |
9.8 |
16 |
Fold:Iron-dependent repressor protein, dimerization domain
Superfamily:Iron-dependent repressor protein, dimerization domain
Family:Iron-dependent repressor protein, dimerization domain |
| 31 | d1sana_ |
|
not modelled |
8.9 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 32 | d1k8ib2 |
|
not modelled |
8.9 |
19 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 33 | d1p7ia_ |
|
not modelled |
8.8 |
31 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 34 | d1f43a_ |
|
not modelled |
8.6 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 35 | c2jobA_ |
|
not modelled |
8.4 |
0 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:antilipopolysaccharide factor;
PDBTitle: solution structure of an antilipopolysaccharide factor from2 shrimp and its possible lipid a binding site
|
| 36 | d1qcva_ |
|
not modelled |
8.2 |
33 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 37 | d6rxna_ |
|
not modelled |
8.2 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 38 | d1wh5a_ |
|
not modelled |
8.1 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 39 | d9anta_ |
|
not modelled |
8.1 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 40 | d1lfba_ |
|
not modelled |
7.9 |
29 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 41 | d1ftza_ |
|
not modelled |
7.9 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 42 | d2dsxa1 |
|
not modelled |
7.9 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 43 | d1yz8p1 |
|
not modelled |
7.9 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 44 | d1jgga_ |
|
not modelled |
7.8 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 45 | d2hrkb1 |
|
not modelled |
7.8 |
30 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Arc1p N-terminal domain-like |
| 46 | d2hddb_ |
|
not modelled |
7.8 |
31 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 47 | c2dmqA_ |
|
not modelled |
7.7 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:lim/homeobox protein lhx9;
PDBTitle: solution structure of the homeobox domain of lim/homeobox2 protein lhx9
|
| 48 | d1ocpa_ |
|
not modelled |
7.7 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 49 | d2o5ha1 |
|
not modelled |
7.6 |
18 |
Fold:NMB0513-like
Superfamily:NMB0513-like
Family:NMB0513-like |
| 50 | d2rdva_ |
|
not modelled |
7.5 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 51 | d1h7va_ |
|
not modelled |
7.4 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 52 | d1brfa_ |
|
not modelled |
7.3 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 53 | d1ijwc_ |
|
not modelled |
7.3 |
32 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |
| 54 | d2craa1 |
|
not modelled |
7.3 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 55 | d1rb9a_ |
|
not modelled |
7.2 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 56 | c2kp6A_ |
|
not modelled |
7.1 |
32 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of protein cv0237 from2 chromobacterium violaceum. northeast structural genomics3 consortium (nesg) target cvt1
|
| 57 | d1iu5a_ |
|
not modelled |
7.1 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 58 | c2da1A_ |
|
not modelled |
7.0 |
31 |
PDB header:transcription
Chain: A: PDB Molecule:alpha-fetoprotein enhancer binding protein;
PDBTitle: solution structure of the first homeobox domain of at-2 binding transcription factor 1 (atbf1)
|
| 59 | d1iroa_ |
|
not modelled |
7.0 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 60 | c1s24A_ |
|
not modelled |
6.9 |
33 |
PDB header:electron transport
Chain: A: PDB Molecule:rubredoxin 2;
PDBTitle: rubredoxin domain ii from pseudomonas oleovorans
|
| 61 | d1s24a_ |
|
not modelled |
6.9 |
33 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 62 | c1x93B_ |
|
not modelled |
6.9 |
40 |
PDB header:transcription
Chain: B: PDB Molecule:hypothetical protein hp0222;
PDBTitle: nmr structure of helicobacter pylori hp0222
|
| 63 | d1x93a1 |
|
not modelled |
6.9 |
40 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 64 | d1fjla_ |
|
not modelled |
6.8 |
38 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 65 | c2dmuA_ |
|
not modelled |
6.8 |
31 |
PDB header:dna binding protein
Chain: A: PDB Molecule:homeobox protein goosecoid;
PDBTitle: solution structure of the homeobox domain of homeobox2 protein goosecoid
|
| 66 | d4rxna_ |
|
not modelled |
6.7 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 67 | c2v3bB_ |
|
not modelled |
6.7 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:rubredoxin 2;
PDBTitle: crystal structure of the electron transfer complex2 rubredoxin - rubredoxin reductase from pseudomonas3 aeruginosa.
|
| 68 | c3fhkF_ |
|
not modelled |
6.7 |
25 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:upf0403 protein yphp;
PDBTitle: crystal structure of apc1446, b.subtilis yphp disulfide2 isomerase
|
| 69 | d1dx8a_ |
|
not modelled |
6.6 |
50 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
| 70 | c2da4A_ |
|
not modelled |
6.6 |
38 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein dkfzp686k21156;
PDBTitle: solution structure of the homeobox domain of the2 hypothetical protein, dkfzp686k21156
|
| 71 | c2l9rA_ |
|
not modelled |
6.5 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:homeobox protein nkx-3.1;
PDBTitle: solution nmr structure of homeobox domain of homeobox protein nkx-3.12 from homo sapiens, northeast structural genomics consortium target3 hr6470a
|
| 72 | d1b8ia_ |
|
not modelled |
6.4 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 73 | d1b72a_ |
|
not modelled |
6.3 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 74 | d1hcra_ |
|
not modelled |
6.3 |
38 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |
| 75 | c2djnA_ |
|
not modelled |
6.1 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:homeobox protein dlx-5;
PDBTitle: the solution structure of the homeobox domain of human2 homeobox protein dlx-5
|
| 76 | d1qasa3 |
|
not modelled |
6.0 |
50 |
Fold:TIM beta/alpha-barrel
Superfamily:PLC-like phosphodiesterases
Family:Mammalian PLC |
| 77 | d1zl8a1 |
|
not modelled |
5.9 |
21 |
Fold:L27 domain
Superfamily:L27 domain
Family:L27 domain |
| 78 | d1vnda_ |
|
not modelled |
5.9 |
44 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 79 | c2dmsA_ |
|
not modelled |
5.9 |
31 |
PDB header:dna binding protein
Chain: A: PDB Molecule:homeobox protein otx2;
PDBTitle: solution structure of the homeobox domain of homeobox2 protein otx2
|
| 80 | c3eq1A_ |
|
not modelled |
5.8 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:porphobilinogen deaminase;
PDBTitle: the crystal structure of human porphobilinogen deaminase at2 2.8a resolution
|
| 81 | c2dmtA_ |
|
not modelled |
5.7 |
25 |
PDB header:dna binding protein
Chain: A: PDB Molecule:homeobox protein barh-like 1;
PDBTitle: solution structure of the homeobox domain of homeobox2 protein barh-like 1
|
| 82 | d2vzsa1 |
|
not modelled |
5.6 |
17 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 83 | d1ztra1 |
|
not modelled |
5.6 |
23 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 84 | c1a31A_ |
|
not modelled |
5.6 |
24 |
PDB header:isomerase/dna
Chain: A: PDB Molecule:protein (topoisomerase i);
PDBTitle: human reconstituted dna topoisomerase i in covalent complex2 with a 22 base pair dna duplex
|
| 85 | c2kn9A_ |
|
not modelled |
5.5 |
33 |
PDB header:electron transport
Chain: A: PDB Molecule:rubredoxin;
PDBTitle: solution structure of zinc-substituted rubredoxin b (rv3250c) from2 mycobacterium tuberculosis. seattle structural genomics center for3 infectious disease target mytud.01635.a
|
| 86 | d1gvda_ |
|
not modelled |
5.5 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
| 87 | c1zcjA_ |
|
not modelled |
5.5 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peroxisomal bifunctional enzyme;
PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase
|
| 88 | c2da2A_ |
|
not modelled |
5.5 |
31 |
PDB header:transcription
Chain: A: PDB Molecule:alpha-fetoprotein enhancer binding protein;
PDBTitle: solution structure of the second homeobox domain of at-2 binding transcription factor 1 (atbf1)
|
| 89 | d1nk3p_ |
|
not modelled |
5.4 |
38 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 90 | d1uhsa_ |
|
not modelled |
5.4 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 91 | d2cufa1 |
|
not modelled |
5.4 |
7 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 92 | c2hfpB_ |
|
not modelled |
5.4 |
83 |
PDB header:transcription
Chain: B: PDB Molecule:src peptide fragment;
PDBTitle: crystal structure of ppar gamma with n-sulfonyl-2-indole2 carboxamide ligands
|
| 93 | c3a01A_ |
|
not modelled |
5.3 |
25 |
PDB header:gene regulation/dna
Chain: A: PDB Molecule:homeodomain-containing protein;
PDBTitle: crystal structure of aristaless and clawless homeodomains bound to dna
|
| 94 | c1k74E_ |
|
not modelled |
5.3 |
83 |
PDB header:transcription
Chain: E: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.3 angstrom resolution crystal structure of the2 heterodimer of the human ppargamma and rxralpha ligand3 binding domains respectively bound with gw409544 and 9-cis4 retinoic acid and co-activator peptides.
|
| 95 | c1fm9E_ |
|
not modelled |
5.3 |
83 |
PDB header:transcription
Chain: E: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.1 angstrom resolution crystal structure of the2 heterodimer of the human rxralpha and ppargamma ligand3 binding domains respectively bound with 9-cis retinoic4 acid and gi262570 and co-activator peptides.
|
| 96 | c1fm6E_ |
|
not modelled |
5.3 |
83 |
PDB header:transcription
Chain: E: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.1 angstrom resolution crystal structure of the2 heterodimer of the human rxralpha and ppargamma ligand3 binding domains respectively bound with 9-cis retinoic4 acid and rosiglitazone and co-activator peptides.
|
| 97 | c3kmgE_ |
|
not modelled |
5.3 |
83 |
PDB header:transcription
Chain: E: PDB Molecule:steroid receptor coactivator-1;
PDBTitle: the x-ray crystal structure of ppar-gamma in complex with an indole2 derivative modulator, gsk538, and an src-1 peptide
|
| 98 | c1fm6V_ |
|
not modelled |
5.3 |
83 |
PDB header:transcription
Chain: V: PDB Molecule:steroid receptor coactivator;
PDBTitle: the 2.1 angstrom resolution crystal structure of the2 heterodimer of the human rxralpha and ppargamma ligand3 binding domains respectively bound with 9-cis retinoic4 acid and rosiglitazone and co-activator peptides.
|
| 99 | d1hdpa_ |
|
not modelled |
5.3 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |