| 1 |
|
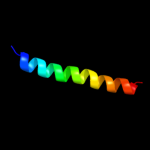
PDB 1m0k chain A
Region: 113 - 166
Aligned: 54
Modelled: 54
Confidence: 14.7%
Identity: 13%
PDB header:ion transport
Chain: A: PDB Molecule:bacteriorhodopsin;
PDBTitle: bacteriorhodopsin k intermediate at 1.43 a resolution
Phyre2
| 2 |
|
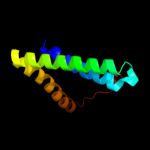
PDB 1m0k chain A
Region: 113 - 166
Aligned: 54
Modelled: 54
Confidence: 14.7%
Identity: 13%
Fold: Family A G protein-coupled receptor-like
Superfamily: Family A G protein-coupled receptor-like
Family: Bacteriorhodopsin-like
Phyre2
| 3 |
|
PDB 1q90 chain B
Region: 73 - 176
Aligned: 95
Modelled: 104
Confidence: 9.9%
Identity: 19%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 4 |
|
PDB 1nek chain C
Region: 75 - 140
Aligned: 63
Modelled: 66
Confidence: 9.3%
Identity: 17%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 5 |
|
PDB 1jb0 chain B
Region: 73 - 130
Aligned: 58
Modelled: 58
Confidence: 9.0%
Identity: 14%
Fold: Photosystem I subunits PsaA/PsaB
Superfamily: Photosystem I subunits PsaA/PsaB
Family: Photosystem I subunits PsaA/PsaB
Phyre2
| 6 |
|
PDB 1jb0 chain A
Region: 73 - 130
Aligned: 58
Modelled: 58
Confidence: 7.6%
Identity: 10%
Fold: Photosystem I subunits PsaA/PsaB
Superfamily: Photosystem I subunits PsaA/PsaB
Family: Photosystem I subunits PsaA/PsaB
Phyre2
| 7 |
|
PDB 2kix chain D
Region: 78 - 97
Aligned: 20
Modelled: 20
Confidence: 7.5%
Identity: 20%
PDB header:transport protein
Chain: D: PDB Molecule:bm2 protein;
PDBTitle: channel domain of bm2 protein from influenza b virus
Phyre2
| 8 |
|
PDB 2e74 chain A domain 1
Region: 73 - 176
Aligned: 95
Modelled: 104
Confidence: 6.8%
Identity: 19%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 9 |
|
PDB 1c8s chain A
Region: 113 - 166
Aligned: 54
Modelled: 54
Confidence: 6.6%
Identity: 13%
Fold: Family A G protein-coupled receptor-like
Superfamily: Family A G protein-coupled receptor-like
Family: Bacteriorhodopsin-like
Phyre2
| 10 |
|
PDB 1xme chain C domain 1
Region: 79 - 88
Aligned: 10
Modelled: 10
Confidence: 6.2%
Identity: 30%
Fold: Single transmembrane helix
Superfamily: Bacterial ba3 type cytochrome c oxidase subunit IIa
Family: Bacterial ba3 type cytochrome c oxidase subunit IIa
Phyre2
| 11 |
|
PDB 1yq3 chain C
Region: 75 - 163
Aligned: 88
Modelled: 89
Confidence: 5.9%
Identity: 13%
PDB header:oxidoreductase
Chain: C: PDB Molecule:succinate dehydrogenase cytochrome b, large subunit;
PDBTitle: avian respiratory complex ii with oxaloacetate and ubiquinone
Phyre2
| 12 |
|
PDB 1v54 chain M
Region: 3 - 32
Aligned: 30
Modelled: 30
Confidence: 5.5%
Identity: 20%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Family: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Phyre2
| 13 |
|
PDB 2ks1 chain B
Region: 114 - 142
Aligned: 29
Modelled: 29
Confidence: 5.5%
Identity: 17%
PDB header:transferase
Chain: B: PDB Molecule:epidermal growth factor receptor;
PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
Phyre2
| 14 |
|
PDB 2l2t chain A
Region: 114 - 142
Aligned: 29
Modelled: 29
Confidence: 5.3%
Identity: 17%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 15 |
|
PDB 3gia chain A
Region: 118 - 208
Aligned: 91
Modelled: 91
Confidence: 5.3%
Identity: 5%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 16 |
|
PDB 2y69 chain Z
Region: 3 - 32
Aligned: 30
Modelled: 30
Confidence: 5.2%
Identity: 20%
PDB header:electron transport
Chain: Z: PDB Molecule:cytochrome c oxidase polypeptide 8h;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2