| 1 |
|
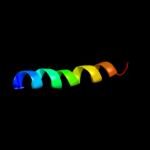
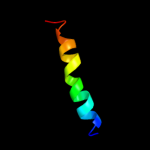
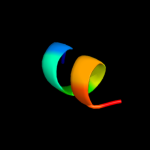
PDB 3lw5 chain 2
Region: 3 - 26
Aligned: 24
Modelled: 24
Confidence: 56.2%
Identity: 21%
PDB header:photosynthesis
Chain: 2: PDB Molecule:type ii chlorophyll a/b binding protein from photosystem i;
PDB Fragment:residues 81-246;
PDBTitle: improved model of plant photosystem i
Phyre2
| 2 |
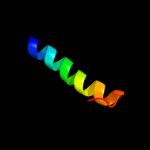
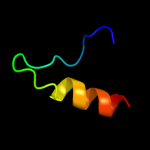
|
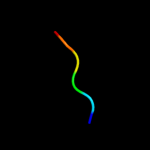
PDB 2wsf chain 2
Region: 3 - 26
Aligned: 24
Modelled: 24
Confidence: 54.2%
Identity: 21%
PDB header:photosynthesis
Chain: 2: PDB Molecule:type ii chlorophyll a/b binding protein from
PDBTitle: improved model of plant photosystem i
Phyre2
| 3 |
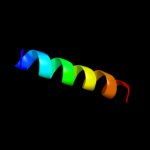
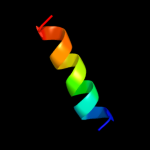
|
PDB 2wse chain 4
Region: 3 - 26
Aligned: 24
Modelled: 24
Confidence: 50.6%
Identity: 29%
PDB header:photosynthesis
Chain: 4: PDB Molecule:chlorophyll a-b binding protein p4,
PDBTitle: improved model of plant photosystem i
Phyre2
| 4 |
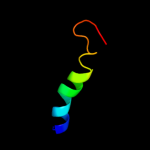
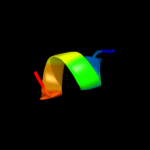
|
PDB 3pl9 chain A
Region: 3 - 26
Aligned: 24
Modelled: 24
Confidence: 48.6%
Identity: 25%
PDB header:photosynthesis
Chain: A: PDB Molecule:chlorophyll a-b binding protein;
PDBTitle: crystal structure of spinach minor light-harvesting complex cp29 at2 2.80 angstrom resolution
Phyre2
| 5 |
|
PDB 1rwt chain A
Region: 3 - 22
Aligned: 20
Modelled: 20
Confidence: 48.3%
Identity: 40%
Fold: Chlorophyll a-b binding protein
Superfamily: Chlorophyll a-b binding protein
Family: Chlorophyll a-b binding protein
Phyre2
| 6 |
|
PDB 2wse chain 3
Region: 3 - 17
Aligned: 15
Modelled: 15
Confidence: 36.3%
Identity: 40%
PDB header:photosynthesis
Chain: 3: PDB Molecule:lhca3;
PDBTitle: improved model of plant photosystem i
Phyre2
| 7 |
|
PDB 2o01 chain 1
Region: 3 - 18
Aligned: 16
Modelled: 16
Confidence: 35.0%
Identity: 44%
PDB header:photosynthesis
Chain: 1: PDB Molecule:at3g54890;
PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
Phyre2
| 8 |
|
PDB 2o01 chain 4
Region: 3 - 26
Aligned: 24
Modelled: 24
Confidence: 34.4%
Identity: 29%
PDB header:photosynthesis
Chain: 4: PDB Molecule:psi light-harvesting antenna chlorophyll a/b-
PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
Phyre2
| 9 |
|
PDB 2o01 chain 2
Region: 3 - 26
Aligned: 24
Modelled: 24
Confidence: 31.9%
Identity: 21%
PDB header:photosynthesis
Chain: 2: PDB Molecule:type ii chlorophyll a/b binding protein from
PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
Phyre2
| 10 |
|
PDB 3brv chain B
Region: 77 - 89
Aligned: 13
Modelled: 13
Confidence: 31.3%
Identity: 38%
PDB header:transferase/transcription
Chain: B: PDB Molecule:nf-kappa-b essential modulator;
PDBTitle: nemo/ikkb association domain structure
Phyre2
| 11 |
|
PDB 1afo chain B
Region: 21 - 45
Aligned: 20
Modelled: 25
Confidence: 31.2%
Identity: 40%
PDB header:integral membrane protein
Chain: B: PDB Molecule:glycophorin a;
PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
Phyre2
| 12 |
|
PDB 2o01 chain 3
Region: 3 - 18
Aligned: 16
Modelled: 16
Confidence: 24.1%
Identity: 44%
PDB header:photosynthesis
Chain: 3: PDB Molecule:psi type iii chlorophyll a/b-binding protein;
PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
Phyre2
| 13 |
|
PDB 2wsc chain 1
Region: 3 - 18
Aligned: 16
Modelled: 16
Confidence: 19.9%
Identity: 44%
PDB header:photosynthesis
Chain: 1: PDB Molecule:at3g54890;
PDBTitle: improved model of plant photosystem i
Phyre2
| 14 |
|
PDB 2q7c chain C
Region: 5 - 15
Aligned: 11
Modelled: 11
Confidence: 12.7%
Identity: 64%
PDB header:viral protein
Chain: C: PDB Molecule:fusion protein between yeast variant gcn4 and
PDBTitle: crystal structure of iqn17
Phyre2
| 15 |
|
PDB 2h3o chain A
Region: 70 - 81
Aligned: 12
Modelled: 12
Confidence: 8.4%
Identity: 42%
PDB header:membrane protein
Chain: A: PDB Molecule:merf;
PDBTitle: structure of merft, a membrane protein with two trans-2 membrane helices
Phyre2
| 16 |
|
PDB 1waz chain A
Region: 70 - 81
Aligned: 12
Modelled: 12
Confidence: 8.1%
Identity: 42%
PDB header:transport protein
Chain: A: PDB Molecule:merf;
PDBTitle: nmr structure determination of the bacterial mercury2 transporter, merf, in micelles
Phyre2
| 17 |
|
PDB 2vxo chain B
Region: 23 - 28
Aligned: 6
Modelled: 6
Confidence: 7.5%
Identity: 83%
PDB header:ligase
Chain: B: PDB Molecule:gmp synthase [glutamine-hydrolyzing];
PDBTitle: human gmp synthetase in complex with xmp
Phyre2
| 18 |
|
PDB 3cyo chain A
Region: 8 - 15
Aligned: 8
Modelled: 8
Confidence: 6.6%
Identity: 75%
PDB header:viral protein
Chain: A: PDB Molecule:transmembrane protein;
PDBTitle: structure of a longer thermalstable core domain of hiv-12 gp41 containing the enfuvirtide resistance mutation n43d3 and complementary mutation e137k
Phyre2
| 19 |
|
PDB 3o3z chain A
Region: 8 - 15
Aligned: 8
Modelled: 8
Confidence: 5.7%
Identity: 75%
PDB header:viral protein
Chain: A: PDB Molecule:envelope glycoprotein gp160;
PDBTitle: complex of a chimeric alpha/beta-peptide based on the gp41 chr domain2 bound to a gp41 nhr domain peptide
Phyre2
| 20 |
|
PDB 3uow chain B
Region: 23 - 28
Aligned: 6
Modelled: 6
Confidence: 5.7%
Identity: 83%
PDB header:ligase
Chain: B: PDB Molecule:gmp synthetase;
PDBTitle: crystal structure of pf10_0123, a gmp synthetase from plasmodium2 falciparum
Phyre2
| 21 |
|