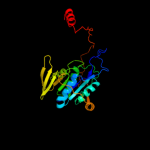
| 1 | d1efvb_ |
|
 |
100.0 |
24 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
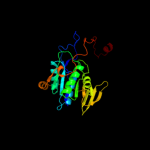
| 2 | d1efpb_ |
|
 |
100.0 |
27 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
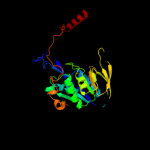
| 3 | d3clsc1 |
|
 |
100.0 |
23 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
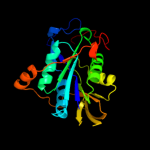
| 4 | d1o94c_ |
|
 |
100.0 |
25 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 5 | c1o94D_ |
|
 |
100.0 |
15 |
PDB header:electron transport
Chain: D: PDB Molecule:electron transfer flavoprotein alpha-subunit;
PDBTitle: ternary complex between trimethylamine dehydrogenase and2 electron transferring flavoprotein
|
| 6 | d3clsd1 |
|
 |
100.0 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 7 | c3ih5A_ |
|
 |
99.9 |
16 |
PDB header:electron transport
Chain: A: PDB Molecule:electron transfer flavoprotein alpha-subunit;
PDBTitle: crystal structure of electron transfer flavoprotein alpha-2 subunit from bacteroides thetaiotaomicron
|
| 8 | d1efva1 |
|
 |
99.9 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 9 | c1t9gR_ |
|
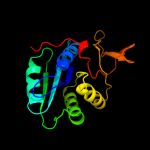 |
99.9 |
17 |
PDB header:oxidoreductase, electron transport
Chain: R: PDB Molecule:electron transfer flavoprotein alpha-subunit,
PDBTitle: structure of the human mcad:etf complex
|
| 10 | c1efpC_ |
|
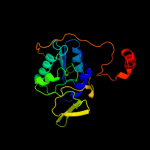 |
99.9 |
15 |
PDB header:electron transport
Chain: C: PDB Molecule:protein (electron transfer flavoprotein);
PDBTitle: electron transfer flavoprotein (etf) from paracoccus2 denitrificans
|
| 11 | d1efpa1 |
|
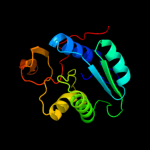 |
99.8 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 12 | c3fetA_ |
|
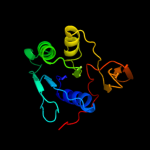 |
99.7 |
11 |
PDB header:electron transport
Chain: A: PDB Molecule:electron transfer flavoprotein subunit alpha related
PDBTitle: crystal structure of the electron transfer flavoprotein subunit alpha2 related protein ta0212 from thermoplasma acidophilum
|
| 13 | d1tq8a_ |
|
 |
95.9 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 14 | c2ixdB_ |
|
 |
95.2 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmbe-related protein;
PDBTitle: crystal structure of the putative deacetylase bc1534 from2 bacilus cereus
|
| 15 | d1uana_ |
|
 |
95.0 |
11 |
Fold:LmbE-like
Superfamily:LmbE-like
Family:LmbE-like |
| 16 | c3fg9B_ |
|
 |
94.2 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein of universal stress protein uspa family;
PDBTitle: the crystal structure of an universal stress protein uspa2 family protein from lactobacillus plantarum wcfs1
|
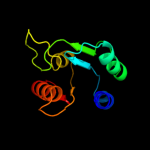
| 17 | c2pfsA_ |
|
 |
94.0 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
|
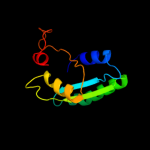
| 18 | c3mt0A_ |
|
 |
93.6 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pa1789;
PDBTitle: the crystal structure of a functionally unknown protein pa1789 from2 pseudomonas aeruginosa pao1
|
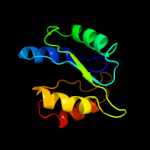
| 19 | c3fh0A_ |
|
 |
93.6 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative universal stress protein kpn_01444;
PDBTitle: crystal structure of putative universal stress protein kpn_01444 -2 atpase
|
| 20 | d1q74a_ |
|
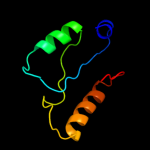 |
92.4 |
15 |
Fold:LmbE-like
Superfamily:LmbE-like
Family:LmbE-like |
| 21 | d2z3va1 |
|
not modelled |
92.0 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 22 | d1q77a_ |
|
not modelled |
90.8 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 23 | c1q7tA_ |
|
not modelled |
89.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein rv1170;
PDBTitle: rv1170 (mshb) from mycobacterium tuberculosis
|
| 24 | c3lpnB_ |
|
not modelled |
89.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of the phosphoribosylpyrophosphate (prpp) synthetase2 from thermoplasma volcanium in complex with an atp analog (ampcpp).
|
| 25 | c3hgmD_ |
|
not modelled |
89.5 |
17 |
PDB header:signaling protein
Chain: D: PDB Molecule:universal stress protein tead;
PDBTitle: universal stress protein tead from the trap transporter2 teaabc of halomonas elongata
|
| 26 | c3loqA_ |
|
not modelled |
88.6 |
14 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: the crystal structure of a universal stress protein from2 archaeoglobus fulgidus dsm 4304
|
| 27 | d1miob_ |
|
not modelled |
88.2 |
15 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 28 | d1m1na_ |
|
not modelled |
84.8 |
13 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 29 | d1m1nb_ |
|
not modelled |
83.6 |
17 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 30 | c3dloC_ |
|
not modelled |
83.5 |
9 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:universal stress protein;
PDBTitle: structure of universal stress protein from archaeoglobus fulgidus
|
| 31 | c1u9yD_ |
|
not modelled |
83.2 |
12 |
PDB header:transferase
Chain: D: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of phosphoribosyl diphosphate synthase2 from methanocaldococcus jannaschii
|
| 32 | d1qh8b_ |
|
not modelled |
82.1 |
14 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 33 | d1ozha1 |
|
not modelled |
81.9 |
11 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 34 | c3dfiA_ |
|
not modelled |
80.8 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:pseudoaglycone deacetylase dbv21;
PDBTitle: the crystal structure of antimicrobial reagent a40926 pseudoaglycone2 deacetylase dbv21
|
| 35 | c1dkrB_ |
|
not modelled |
80.3 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosyl pyrophosphate synthetase;
PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
|
| 36 | c3pdiG_ |
|
not modelled |
77.6 |
12 |
PDB header:protein binding
Chain: G: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nife;
PDBTitle: precursor bound nifen
|
| 37 | d2gm3a1 |
|
not modelled |
77.4 |
11 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 38 | d1f6da_ |
|
not modelled |
77.2 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
| 39 | d2ji7a1 |
|
not modelled |
76.9 |
12 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 40 | d2iyva1 |
|
not modelled |
76.5 |
10 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 41 | c2dy0A_ |
|
not modelled |
75.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:adenine phosphoribosyltransferase;
PDBTitle: crystal structure of project jw0458 from escherichia coli
|
| 42 | d1qh8a_ |
|
not modelled |
74.7 |
16 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 43 | d1a9xa3 |
|
not modelled |
73.5 |
16 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 44 | d1a9xa4 |
|
not modelled |
73.1 |
19 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 45 | d1jmva_ |
|
not modelled |
72.8 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 46 | c2ebdB_ |
|
not modelled |
71.9 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] synthase 3;
PDBTitle: crystal structure of 3-oxoacyl-[acyl-carrier-protein] synthase iii2 from aquifex aeolicus vf5
|
| 47 | d1o57a2 |
|
not modelled |
71.8 |
17 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 48 | c3s3tD_ |
|
not modelled |
71.4 |
17 |
PDB header:chaperone
Chain: D: PDB Molecule:nucleotide-binding protein, universal stress protein uspa
PDBTitle: universal stress protein uspa from lactobacillus plantarum
|
| 49 | d1rkba_ |
|
not modelled |
70.4 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 50 | d1l1qa_ |
|
not modelled |
69.6 |
12 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 51 | d1y0ba1 |
|
not modelled |
69.3 |
17 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 52 | d1v4va_ |
|
not modelled |
69.2 |
21 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
| 53 | d1kaga_ |
|
not modelled |
69.2 |
9 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 54 | c3il5D_ |
|
not modelled |
68.9 |
11 |
PDB header:transferase
Chain: D: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] synthase 3;
PDBTitle: structure of e. faecalis fabh in complex with 2-({4-bromo-3-2 [(diethylamino)sulfonyl]benzoyl}amino)benzoic acid
|
| 55 | d1iowa1 |
|
not modelled |
68.0 |
20 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:D-Alanine ligase N-terminal domain |
| 56 | c2c4kD_ |
|
not modelled |
67.1 |
11 |
PDB header:regulatory protein
Chain: D: PDB Molecule:phosphoribosyl pyrophosphate synthetase-
PDBTitle: crystal structure of human phosphoribosylpyrophosphate2 synthetase-associated protein 39 (pap39)
|
| 57 | d1g2qa_ |
|
not modelled |
67.0 |
15 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 58 | d1o6ca_ |
|
not modelled |
66.8 |
13 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
| 59 | c2qv5A_ |
|
not modelled |
66.6 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu2773;
PDBTitle: crystal structure of uncharacterized protein atu2773 from2 agrobacterium tumefaciens c58
|
| 60 | c3efhB_ |
|
not modelled |
65.5 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase 1;
PDBTitle: crystal structure of human phosphoribosyl pyrophosphate2 synthetase 1
|
| 61 | c3olqA_ |
|
not modelled |
64.4 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:universal stress protein e;
PDBTitle: the crystal structure of a universal stress protein e from proteus2 mirabilis hi4320
|
| 62 | c2ip4A_ |
|
not modelled |
63.5 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of glycinamide ribonucleotide synthetase from2 thermus thermophilus hb8
|
| 63 | c1zuiA_ |
|
not modelled |
61.6 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:shikimate kinase;
PDBTitle: structural basis for shikimate-binding specificity of helicobacter2 pylori shikimate kinase
|
| 64 | c3ab8B_ |
|
not modelled |
61.6 |
20 |
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein ttha0350;
PDBTitle: crystal structure of the hypothetical tandem-type universal stress2 protein ttha0350 complexed with atps.
|
| 65 | c2dumD_ |
|
not modelled |
61.5 |
18 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein ph0823;
PDBTitle: crystal structure of hypothetical protein, ph0823
|
| 66 | d2djia1 |
|
not modelled |
61.0 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 67 | d2ez9a1 |
|
not modelled |
60.0 |
13 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 68 | c3ot5D_ |
|
not modelled |
59.9 |
10 |
PDB header:isomerase
Chain: D: PDB Molecule:udp-n-acetylglucosamine 2-epimerase;
PDBTitle: 2.2 angstrom resolution crystal structure of putative udp-n-2 acetylglucosamine 2-epimerase from listeria monocytogenes
|
| 69 | d1ybha1 |
|
not modelled |
59.5 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 70 | c1kbiB_ |
|
not modelled |
58.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome b2;
PDBTitle: crystallographic study of the recombinant flavin-binding domain of2 baker's yeast flavocytochrome b2: comparison with the intact wild-3 type enzyme
|
| 71 | c1o57A_ |
|
not modelled |
58.8 |
14 |
PDB header:dna binding protein
Chain: A: PDB Molecule:pur operon repressor;
PDBTitle: crystal structure of the purine operon repressor of2 bacillus subtilis
|
| 72 | c2nz2A_ |
|
not modelled |
58.5 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of human argininosuccinate synthase in complex with2 aspartate and citrulline
|
| 73 | d2ptza1 |
|
not modelled |
58.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 74 | d1viaa_ |
|
not modelled |
57.6 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 75 | d1e6ca_ |
|
not modelled |
57.6 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 76 | c3dfmA_ |
|
not modelled |
57.5 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:teicoplanin pseudoaglycone deacetylase orf2;
PDBTitle: the crystal structure of the zinc inhibited form of2 teicoplanin deacetylase orf2
|
| 77 | c1w96B_ |
|
not modelled |
57.0 |
10 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase;
PDBTitle: crystal structure of biotin carboxylase domain of acetyl-2 coenzyme a carboxylase from saccharomyces cerevisiae in3 complex with soraphen a
|
| 78 | c3s29C_ |
|
not modelled |
56.8 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:sucrose synthase 1;
PDBTitle: the crystal structure of sucrose synthase-1 from arabidopsis thaliana2 and its functional implications.
|
| 79 | c3op1A_ |
|
not modelled |
56.6 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:macrolide-efflux protein;
PDBTitle: crystal structure of macrolide-efflux protein sp_1110 from2 streptococcus pneumoniae
|
| 80 | d1vcfa1 |
|
not modelled |
56.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 81 | d1w0ma_ |
|
not modelled |
56.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 82 | c2pt5D_ |
|
not modelled |
56.3 |
22 |
PDB header:transferase
Chain: D: PDB Molecule:shikimate kinase;
PDBTitle: crystal structure of shikimate kinase (aq_2177) from aquifex aeolicus2 vf5
|
| 83 | d1jlja_ |
|
not modelled |
55.3 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 84 | c3t61A_ |
|
not modelled |
54.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:gluconokinase;
PDBTitle: crystal structure of a gluconokinase from sinorhizobium meliloti 1021
|
| 85 | d1s3ga1 |
|
not modelled |
54.7 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 86 | c3fdiA_ |
|
not modelled |
54.3 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein from eubacterium2 ventriosum atcc 27560.
|
| 87 | d2cdna1 |
|
not modelled |
53.7 |
21 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 88 | d1qb7a_ |
|
not modelled |
53.2 |
14 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 89 | c1kh2D_ |
|
not modelled |
52.4 |
19 |
PDB header:ligase
Chain: D: PDB Molecule:argininosuccinate synthetase;
PDBTitle: crystal structure of thermus thermophilus hb82 argininosuccinate synthetase in complex with atp
|
| 90 | d1ovma1 |
|
not modelled |
52.4 |
12 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 91 | d1zpda1 |
|
not modelled |
52.3 |
13 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 92 | c2v4wB_ |
|
not modelled |
52.0 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:hydroxymethylglutaryl-coa synthase,
PDBTitle: crystal structure of human mitochondrial 3-hydroxy-3-2 methylglutaryl-coenzyme a synthase 2 (hmgcs2)
|
| 93 | c3jyfB_ |
|
not modelled |
51.7 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-
PDBTitle: the crystal structure of a 2,3-cyclic nucleotide 2-2 phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor3 protein from klebsiella pneumoniae subsp. pneumoniae mgh 78578
|
| 94 | c2h92C_ |
|
not modelled |
51.2 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:cytidylate kinase;
PDBTitle: crystal structure of staphylococcus aureus cytidine2 monophosphate kinase in complex with cytidine-5'-3 monophosphate
|
| 95 | c2vu2D_ |
|
not modelled |
50.8 |
11 |
PDB header:transferase
Chain: D: PDB Molecule:acetyl-coa acetyltransferase;
PDBTitle: biosynthetic thiolase from z. ramigera. complex with s-2 pantetheine-11-pivalate.
|
| 96 | c3gwaA_ |
|
not modelled |
50.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) synthase iii;
PDBTitle: 1.6 angstrom crystal structure of 3-oxoacyl-(acyl-carrier-protein)2 synthase iii
|
| 97 | c3gmtB_ |
|
not modelled |
50.8 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:adenylate kinase;
PDBTitle: crystal structure of adenylate kinase from burkholderia pseudomallei
|
| 98 | d1zina1 |
|
not modelled |
50.5 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 99 | c2eu8B_ |
|
not modelled |
50.3 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:adenylate kinase;
PDBTitle: crystal structure of a thermostable mutant of bacillus2 subtilis adenylate kinase (q199r)
|
| 100 | c3hdtB_ |
|
not modelled |
50.1 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative kinase;
PDBTitle: crystal structure of putative kinase from clostridium symbiosum atcc2 14940
|
| 101 | c1m6vE_ |
|
not modelled |
50.0 |
16 |
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
|
| 102 | c2bwjC_ |
|
not modelled |
49.7 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:adenylate kinase 5;
PDBTitle: structure of adenylate kinase 5
|
| 103 | c2grjH_ |
|
not modelled |
49.6 |
14 |
PDB header:transferase
Chain: H: PDB Molecule:dephospho-coa kinase;
PDBTitle: crystal structure of dephospho-coa kinase (ec 2.7.1.24)2 (dephosphocoenzyme a kinase) (tm1387) from thermotoga maritima at3 2.60 a resolution
|
| 104 | d1di6a_ |
|
not modelled |
49.5 |
10 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 105 | c2ar7A_ |
|
not modelled |
49.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate kinase 4;
PDBTitle: crystal structure of human adenylate kinase 4, ak4
|
| 106 | d1e4va1 |
|
not modelled |
49.3 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 107 | d1vcha1 |
|
not modelled |
48.7 |
22 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 108 | c1ankA_ |
|
not modelled |
48.4 |
14 |
PDB header:transferase(phosphotransferase)
Chain: A: PDB Molecule:adenylate kinase;
PDBTitle: the closed conformation of a highly flexible protein: the2 structure of e. coli adenylate kinase with bound amp and3 amppnp
|
| 109 | c3be4A_ |
|
not modelled |
48.1 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate kinase;
PDBTitle: crystal structure of cryptosporidium parvum adenylate kinase cgd5_3360
|
| 110 | d1mzva_ |
|
not modelled |
48.0 |
14 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 111 | d1zaka1 |
|
not modelled |
48.0 |
21 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 112 | d1rqba2 |
|
not modelled |
47.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 113 | c2rhmD_ |
|
not modelled |
47.9 |
14 |
PDB header:unknown function
Chain: D: PDB Molecule:putative kinase;
PDBTitle: crystal structure of a putative kinase (caur_3907) from chloroflexus2 aurantiacus j-10-fl at 1.70 a resolution
|
| 114 | d1knqa_ |
|
not modelled |
47.9 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Gluconate kinase |
| 115 | d1zn7a1 |
|
not modelled |
47.6 |
20 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 116 | d2ihta1 |
|
not modelled |
47.5 |
23 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 117 | c3r8cB_ |
|
not modelled |
47.0 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:cytidylate kinase;
PDBTitle: crystal structure of cytidylate kinase (cmk) from mycobacterium2 abscessus
|
| 118 | c2vliB_ |
|
not modelled |
46.8 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:antibiotic resistance protein;
PDBTitle: structure of deinococcus radiodurans tunicamycin resistance2 protein
|
| 119 | c1s3gA_ |
|
not modelled |
46.2 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:adenylate kinase;
PDBTitle: crystal structure of adenylate kinase from bacillus2 globisporus
|
| 120 | c3trfB_ |
|
not modelled |
45.8 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:shikimate kinase;
PDBTitle: structure of a shikimate kinase (arok) from coxiella burnetii
|

