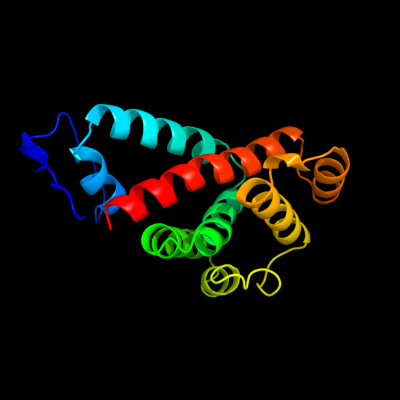
1 d2prra1
100.0
16
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: Atu0492-like2 d2pfxa1
100.0
13
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: Atu0492-like3 c3c1lB_
100.0
15
PDB header: oxidoreductaseChain: B: PDB Molecule: putative antioxidant defense protein mlr4105;PDBTitle: crystal structure of an antioxidant defense protein (mlr4105) from2 mesorhizobium loti maff303099 at 2.00 a resolution
4 c3lvyB_
100.0
14
PDB header: lyaseChain: B: PDB Molecule: carboxymuconolactone decarboxylase family;PDBTitle: crystal structure of carboxymuconolactone decarboxylase2 family protein smu.961 from streptococcus mutans
5 d2oyoa1
100.0
19
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: Atu0492-like6 d2o4da1
100.0
16
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: Atu0492-like7 d2gmya1
100.0
16
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: Atu0492-like8 d2ouwa1
99.7
18
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: TTHA0727-like9 c3beyC_
99.0
12
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: conserved protein o27018;PDBTitle: crystal structure of the protein o27018 from methanobacterium2 thermoautotrophicum. northeast structural genomics consortium target3 tt217
10 d1vkea_
98.9
11
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like11 c1p8cD_
98.9
10
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of tm1620 (apc4843) from thermotoga2 maritima
12 c2qeuA_
98.9
12
PDB header: lyaseChain: A: PDB Molecule: putative carboxymuconolactone decarboxylase;PDBTitle: crystal structure of putative carboxymuconolactone decarboxylase2 (yp_555818.1) from burkholderia xenovorans lb400 at 1.65 a resolution
13 d1knca_
98.8
13
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: AhpD14 c3d7iB_
98.6
12
PDB header: lyaseChain: B: PDB Molecule: carboxymuconolactone decarboxylase family protein;PDBTitle: crystal structure of carboxymuconolactone decarboxylase family protein2 possibly involved in oxygen detoxification (1591455) from3 methanococcus jannaschii at 1.75 a resolution
15 d2q0ta1
98.5
12
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: AhpD16 d2cwqa1
98.4
16
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: TTHA0727-like17 d1vkeb_
98.3
7
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like18 d2af7a1
96.6
7
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like19 d1a9xa1
92.1
9
Fold: Carbamoyl phosphate synthetase, large subunit connection domainSuperfamily: Carbamoyl phosphate synthetase, large subunit connection domainFamily: Carbamoyl phosphate synthetase, large subunit connection domain20 c3bjxB_
68.5
13
PDB header: hydrolaseChain: B: PDB Molecule: halocarboxylic acid dehalogenase dehi;PDBTitle: structure of a group i haloacid dehalogenase from2 pseudomonas putida strain pp3
21 c3cixA_
not modelled
54.4
11
PDB header: adomet binding proteinChain: A: PDB Molecule: fefe-hydrogenase maturase;PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
22 c3iwfA_
not modelled
41.4
16
PDB header: transcription regulatorChain: A: PDB Molecule: transcription regulator rpir family;PDBTitle: the crystal structure of the n-terminal domain of a rpir2 transcriptional regulator from staphylococcus epidermidis to 1.4a
23 d2a1ka1
not modelled
39.9
43
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Phage ssDNA-binding proteins24 c3omdB_
not modelled
39.4
7
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of unknown function protein from leptospirillum2 rubarum
25 d1bkra_
not modelled
38.9
25
Fold: CH domain-likeSuperfamily: Calponin-homology domain, CH-domainFamily: Calponin-homology domain, CH-domain26 c2d87A_
not modelled
38.2
15
PDB header: structural protein, protein bindingChain: A: PDB Molecule: smoothelin splice isoform l2;PDBTitle: solution structure of the ch domain from human smoothelin2 splice isoform l2
27 c1m6vE_
not modelled
36.3
9
PDB header: ligaseChain: E: PDB Molecule: carbamoyl phosphate synthetase large chain;PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
28 c2vn2B_
not modelled
35.9
20
PDB header: replicationChain: B: PDB Molecule: chromosome replication initiation protein;PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
29 d1gpca_
not modelled
33.7
43
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Phage ssDNA-binding proteins30 c2d89A_
not modelled
31.6
18
PDB header: structural protein, protein bindingChain: A: PDB Molecule: ehbp1 protein;PDBTitle: solution structure of the ch domain from human eh domain2 binding protein 1
31 c2krkA_
not modelled
31.4
16
PDB header: protein bindingChain: A: PDB Molecule: 26s protease regulatory subunit 8;PDBTitle: solution nmr structure of 26s protease regulatory subunit 82 from h.sapiens, northeast structural genomics consortium3 target target hr3102a
32 c1wylA_
not modelled
30.5
30
PDB header: signaling proteinChain: A: PDB Molecule: nedd9 interacting protein with calponin homologyPDBTitle: solution structure of the ch domain of human nedd92 interacting protein with calponin homology and lim domains
33 c3mhvC_
not modelled
28.6
13
PDB header: protein transportChain: C: PDB Molecule: vacuolar protein sorting-associated protein 4;PDBTitle: crystal structure of vps4 and vta1
34 c3rmsA_
not modelled
26.8
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein svir_20580 from2 saccharomonospora viridis
35 d1t3wa_
not modelled
24.7
15
Fold: DNA primase DnaG, C-terminal domainSuperfamily: DNA primase DnaG, C-terminal domainFamily: DNA primase DnaG, C-terminal domain36 d2csba4
not modelled
23.0
29
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Topoisomerase V repeat domain37 d1t33a2
not modelled
22.9
10
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain38 c1wr1B_
not modelled
22.7
20
PDB header: signaling proteinChain: B: PDB Molecule: ubiquitin-like protein dsk2;PDBTitle: the complex sturcture of dsk2p uba with ubiquitin
39 c2dt7A_
not modelled
20.5
36
PDB header: rna binding proteinChain: A: PDB Molecule: splicing factor 3a subunit 3;PDBTitle: solution structure of the second surp domain of human2 splicing factor sf3a120 in complex with a fragment of3 human splicing factor sf3a60
40 c2v79B_
not modelled
20.3
20
PDB header: dna-binding proteinChain: B: PDB Molecule: dna replication protein dnad;PDBTitle: crystal structure of the n-terminal domain of dnad from2 bacillus subtilis
41 c1t3bA_
not modelled
20.2
11
PDB header: isomeraseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbc;PDBTitle: x-ray structure of dsbc from haemophilus influenzae
42 c3n7qA_
not modelled
19.1
19
PDB header: transcription, replication/dnaChain: A: PDB Molecule: transcription termination factor, mitochondrial;PDBTitle: crystal structure of human mitochondrial mterf fragment (aa 99-399) in2 complex with a 12-mer dna encompassing the trnaleu(uur) binding3 sequence
43 d1sh5a2
not modelled
18.8
28
Fold: CH domain-likeSuperfamily: Calponin-homology domain, CH-domainFamily: Calponin-homology domain, CH-domain44 c3l9vE_
not modelled
18.7
33
PDB header: oxidoreductaseChain: E: PDB Molecule: putative thiol-disulfide isomerase or thioredoxin;PDBTitle: crystal structure of salmonella enterica serovar typhimurium srga
45 c3dvwA_
not modelled
18.5
22
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsba;PDBTitle: crystal structure of reduced dsba1 from neisseria2 meningitidis
46 c2rcyB_
not modelled
18.1
12
PDB header: oxidoreductaseChain: B: PDB Molecule: pyrroline carboxylate reductase;PDBTitle: crystal structure of plasmodium falciparum pyrroline carboxylate2 reductase (mal13p1.284) with nadp bound
47 c2d88A_
not modelled
18.0
13
PDB header: signaling protein, protein bindingChain: A: PDB Molecule: protein mical-3;PDBTitle: solution structure of the ch domain from human mical-32 protein
48 d1bhda_
not modelled
17.5
25
Fold: CH domain-likeSuperfamily: Calponin-homology domain, CH-domainFamily: Calponin-homology domain, CH-domain49 c3bj4B_
not modelled
17.0
41
PDB header: signaling proteinChain: B: PDB Molecule: potassium voltage-gated channel subfamily kqtPDBTitle: the kcnq1 (kv7.1) c-terminus, a multi-tiered scaffold for2 subunit assembly and protein interaction
50 c3lgcA_
not modelled
16.9
25
PDB header: unknown functionChain: A: PDB Molecule: glutaredoxin 1;PDBTitle: crystal structure of glutaredoxin 1 from francisella2 tularensis
51 d1sxjd1
not modelled
15.8
11
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: DNA polymerase III clamp loader subunits, C-terminal domain52 c1v57A_
not modelled
15.8
18
PDB header: isomeraseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbg;PDBTitle: crystal structure of the disulfide bond isomerase dsbg
53 c3hd5A_
not modelled
15.1
11
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsba;PDBTitle: crystal structure of a thiol:disulfide interchange protein2 dsba from bordetella parapertussis
54 c3gv1A_
not modelled
15.0
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: disulfide interchange protein;PDBTitle: crystal structure of disulfide interchange protein from neisseria2 gonorrhoeae
55 c2jy5A_
not modelled
14.7
13
PDB header: signaling proteinChain: A: PDB Molecule: ubiquilin-1;PDBTitle: nmr structure of ubiquilin 1 uba domain
56 d2ccaa1
not modelled
14.5
11
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG57 d1xcca_
not modelled
14.5
25
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like58 d1t3ba1
not modelled
14.3
10
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbC/DsbG C-terminal domain-like59 c2remB_
not modelled
13.8
22
PDB header: oxidoreductaseChain: B: PDB Molecule: disulfide oxidoreductase;PDBTitle: crystal structure of oxidoreductase dsba from xylella2 fastidiosa
60 d1m3va1
not modelled
13.7
56
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain61 d1rt8a_
not modelled
13.7
23
Fold: CH domain-likeSuperfamily: Calponin-homology domain, CH-domainFamily: Calponin-homology domain, CH-domain62 c3hi2D_
not modelled
13.6
11
PDB header: dna binding protein/toxinChain: D: PDB Molecule: motility quorum-sensing regulator mqsr;PDBTitle: structure of the n-terminal domain of the e. coli antitoxin mqsa2 (ygit/b3021) in complex with the e. coli toxin mqsr (ygiu/b3022)
63 c3gykC_
not modelled
13.1
14
PDB header: oxidoreductaseChain: C: PDB Molecule: 27kda outer membrane protein;PDBTitle: the crystal structure of a thioredoxin-like oxidoreductase from2 silicibacter pomeroyi dss-3
64 c1jzdA_
not modelled
13.0
0
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbc;PDBTitle: dsbc-dsbdalpha complex
65 c3riwA_
not modelled
12.8
15
PDB header: oxidoreductaseChain: A: PDB Molecule: ascorbate peroxidase;PDBTitle: the crystal structure of leishmania major peroxidase mutant c197t
66 d1eeja1
not modelled
12.3
11
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbC/DsbG C-terminal domain-like67 c2k9hA_
not modelled
12.1
35
PDB header: metal binding proteinChain: A: PDB Molecule: glycoprotein;PDBTitle: the hantavirus glycoprotein g1 tail contains a dual cchc-2 type classical zinc fingers
68 d2vkva2
not modelled
11.8
10
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain69 c3ghaA_
not modelled
11.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: disulfide bond formation protein d;PDBTitle: crystal structure of etda-treated bdbd (reduced)
70 c3c7mB_
not modelled
11.6
13
PDB header: oxidoreductaseChain: B: PDB Molecule: thiol:disulfide interchange protein dsba-like;PDBTitle: crystal structure of reduced dsbl
71 d1prxa_
not modelled
11.4
25
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like72 d2o3fa1
not modelled
11.4
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: RpiR-like73 c3feuA_
not modelled
11.3
11
PDB header: oxidoreductaseChain: A: PDB Molecule: putative lipoprotein;PDBTitle: crystal structure of dsba-like thioredoxin domain vf_a0457 from vibrio2 fischeri
74 d2euta1
not modelled
11.2
7
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: CCP-like75 c2v2gC_
not modelled
11.2
29
PDB header: oxidoreductaseChain: C: PDB Molecule: peroxiredoxin 6;PDBTitle: crystal structure of the c45s mutant of the peroxiredoxin 62 of arenicola marina. monoclinic form
76 c2qjxA_
not modelled
11.0
18
PDB header: protein bindingChain: A: PDB Molecule: protein bim1;PDBTitle: structural basis of microtubule plus end tracking by2 xmap215, clip-170 and eb1
77 d1repc1
not modelled
10.7
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Replication initiation protein78 d1pxya_
not modelled
10.6
19
Fold: CH domain-likeSuperfamily: Calponin-homology domain, CH-domainFamily: Calponin-homology domain, CH-domain79 d1j2oa1
not modelled
10.5
22
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain80 c3mk7B_
not modelled
10.5
33
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit o;PDBTitle: the structure of cbb3 cytochrome oxidase
81 c3bciA_
not modelled
10.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: disulfide bond protein a;PDBTitle: crystal structure of staphylococcus aureus dsba
82 d2zcta1
not modelled
10.3
15
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like83 c2o3fC_
not modelled
10.1
17
PDB header: transcriptionChain: C: PDB Molecule: putative hth-type transcriptional regulator ybbh;PDBTitle: structural genomics, the crystal structure of the n-2 terminal domain of the putative transcriptional regulator3 ybbh from bacillus subtilis subsp. subtilis str. 168.
84 d1un2a_
not modelled
10.1
22
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbA-like85 d1ehsa_
not modelled
10.0
50
Fold: Toxic hairpinSuperfamily: Heat-stable enterotoxin BFamily: Heat-stable enterotoxin B86 d1dxxa2
not modelled
9.8
25
Fold: CH domain-likeSuperfamily: Calponin-homology domain, CH-domainFamily: Calponin-homology domain, CH-domain87 c3gmfA_
not modelled
9.8
20
PDB header: oxidoreductaseChain: A: PDB Molecule: protein-disulfide isomerase;PDBTitle: crystal structure of protein-disulfide isomerase from novosphingobium2 aromaticivorans
88 d2daha1
not modelled
9.1
13
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain89 d2e39a1
not modelled
9.0
15
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: CCP-like90 d1oqya2
not modelled
8.9
9
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain91 d1rutx1
not modelled
8.9
63
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain92 d2bwba1
not modelled
8.9
14
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain93 d1h75a_
not modelled
8.7
31
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase94 c1g6uB_
not modelled
8.7
15
PDB header: de novo proteinChain: B: PDB Molecule: domain swapped dimer;PDBTitle: crystal structure of a domain swapped dimer
95 d1mwva1
not modelled
8.7
15
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Catalase-peroxidase KatG96 d1fova_
not modelled
8.6
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase97 c3nznA_
not modelled
8.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin;PDBTitle: the crystal structure of the glutaredoxin from methanosarcina mazei2 go1
98 c1nm3B_
not modelled
8.4
23
PDB header: electron transportChain: B: PDB Molecule: protein hi0572;PDBTitle: crystal structure of heamophilus influenza hybrid-prx5
99 c2agaA_
not modelled
8.3
33
PDB header: transcriptionChain: A: PDB Molecule: machado-joseph disease protein 1;PDBTitle: de-ubiquitinating function of ataxin-3: insights from the2 solution structure of the josephin domain