| 1 |
|
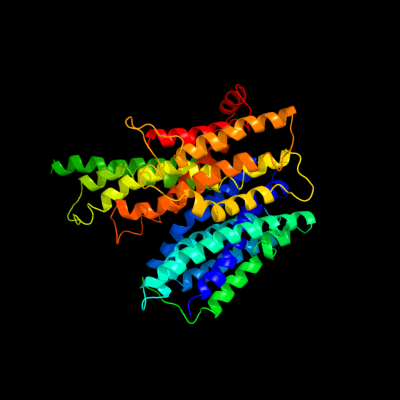
PDB 2xut chain C
Region: 10 - 472
Aligned: 448
Modelled: 463
Confidence: 100.0%
Identity: 18%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 2 |
|
PDB 1pw4 chain A
Region: 2 - 492
Aligned: 412
Modelled: 427
Confidence: 100.0%
Identity: 14%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 3 |
|
PDB 1pv7 chain A
Region: 9 - 492
Aligned: 409
Modelled: 411
Confidence: 100.0%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 4 |
|
PDB 3o7p chain A
Region: 12 - 480
Aligned: 405
Modelled: 408
Confidence: 100.0%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 5 |
|
PDB 2gfp chain A
Region: 11 - 478
Aligned: 372
Modelled: 379
Confidence: 99.9%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 6 |
|
PDB 1ikp chain A domain 3
Region: 3 - 46
Aligned: 37
Modelled: 38
Confidence: 12.4%
Identity: 30%
Fold: Toxins' membrane translocation domains
Superfamily: Exotoxin A, middle domain
Family: Exotoxin A, middle domain
Phyre2
| 7 |
|
PDB 1fft chain B domain 2
Region: 439 - 488
Aligned: 50
Modelled: 50
Confidence: 10.8%
Identity: 14%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 8 |
|
PDB 3hzq chain A
Region: 412 - 488
Aligned: 77
Modelled: 77
Confidence: 7.5%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: structure of a tetrameric mscl in an expanded intermediate2 state
Phyre2
| 9 |
|
PDB 1j4n chain A
Region: 160 - 203
Aligned: 44
Modelled: 44
Confidence: 7.1%
Identity: 7%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 10 |
|
PDB 2knc chain A
Region: 458 - 493
Aligned: 36
Modelled: 36
Confidence: 6.8%
Identity: 25%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 11 |
|
PDB 3pro chain C domain 1
Region: 30 - 74
Aligned: 45
Modelled: 45
Confidence: 6.8%
Identity: 9%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Alpha-lytic protease prodomain
Family: Alpha-lytic protease prodomain
Phyre2
| 12 |
|
PDB 2g9p chain A
Region: 64 - 78
Aligned: 14
Modelled: 15
Confidence: 6.0%
Identity: 36%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 13 |
|
PDB 1ikq chain A
Region: 3 - 47
Aligned: 38
Modelled: 45
Confidence: 6.0%
Identity: 29%
PDB header:transferase
Chain: A: PDB Molecule:exotoxin a;
PDBTitle: pseudomonas aeruginosa exotoxin a, wild type
Phyre2
| 14 |
|
PDB 1o82 chain A
Region: 36 - 69
Aligned: 34
Modelled: 34
Confidence: 5.9%
Identity: 15%
Fold: Saposin-like
Superfamily: Bacteriocin AS-48
Family: Bacteriocin AS-48
Phyre2
| 15 |
|
PDB 2rdd chain B
Region: 461 - 493
Aligned: 33
Modelled: 33
Confidence: 5.6%
Identity: 15%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 16 |
|
PDB 2ox6 chain A domain 1
Region: 27 - 51
Aligned: 25
Modelled: 25
Confidence: 5.5%
Identity: 24%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: YdiL-like
Phyre2
| 17 |
|
PDB 2hoe chain A domain 1
Region: 36 - 54
Aligned: 19
Modelled: 19
Confidence: 5.1%
Identity: 21%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: ROK associated domain
Phyre2
| 18 |
|
PDB 2auw chain A domain 1
Region: 2 - 16
Aligned: 15
Modelled: 15
Confidence: 5.1%
Identity: 27%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: NE0471 C-terminal domain-like
Phyre2