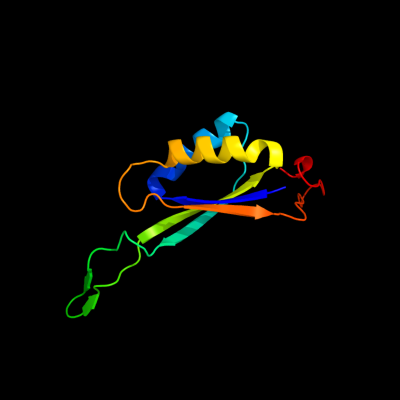
| 1 | d2ns1b1
|
|
 |
100.0 |
99 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
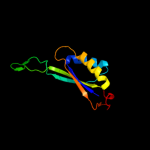
| 2 | c3mhyC_
|
|
 |
100.0 |
68 |
PDB header:signaling protein
Chain: C: PDB Molecule:pii-like protein pz;
PDBTitle: a new pii protein structure
|
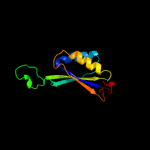
| 3 | d1qy7a_
|
|
 |
100.0 |
56 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
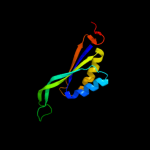
| 4 | d2piia_
|
|
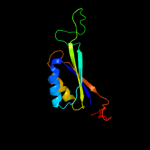 |
100.0 |
68 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 5 | d1vfja_
|
|
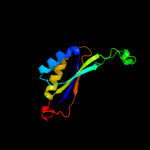 |
100.0 |
40 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 6 | c2rd5D_
|
|
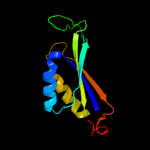 |
100.0 |
45 |
PDB header:protein binding
Chain: D: PDB Molecule:pii protein;
PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
|
| 7 | c3ncpD_
|
|
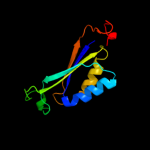 |
100.0 |
46 |
PDB header:signaling protein
Chain: D: PDB Molecule:nitrogen regulatory protein p-ii (glnb-2);
PDBTitle: glnk2 from archaeoglobus fulgidus
|
| 8 | d1hwua_
|
|
 |
100.0 |
63 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 9 | d1ul3a_
|
|
 |
100.0 |
55 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 10 | c2j9dG_
|
|
 |
100.0 |
53 |
PDB header:membrane transport
Chain: G: PDB Molecule:hypothetical nitrogen regulatory pii-like
PDBTitle: structure of glnk1 with bound effectors indicates2 regulatory mechanism for ammonia uptake
|
| 11 | c3bzqA_
|
|
 |
100.0 |
49 |
PDB header:signaling protein/transcription
Chain: A: PDB Molecule:nitrogen regulatory protein p-ii;
PDBTitle: high resolution crystal structure of nitrogen regulatory protein2 (rv2919c) of mycobacterium tuberculosis
|
| 12 | c3o8wA_
|
|
 |
100.0 |
50 |
PDB header:signaling protein
Chain: A: PDB Molecule:nitrogen regulatory protein p-ii (glnb-1);
PDBTitle: archaeoglobus fulgidus glnk1
|
| 13 | c3l7pA_
|
|
 |
99.9 |
48 |
PDB header:transcription
Chain: A: PDB Molecule:putative nitrogen regulatory protein pii;
PDBTitle: crystal structure of smu.1657c, putative nitrogen regulatory protein2 pii from streptococcus mutans
|
| 14 | d2cz4a1
|
|
 |
99.8 |
17 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 15 | c3ce8A_
|
|
 |
98.5 |
14 |
PDB header:unknown function
Chain: A: PDB Molecule:putative pii-like nitrogen regulatory protein;
PDBTitle: crystal structure of a duf3240 family protein (sbal_0098) from2 shewanella baltica os155 at 2.40 a resolution
|
| 16 | c3m05A_
|
|
 |
98.2 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pepe_1480;
PDBTitle: the crystal structure of a functionally unknown protein2 pepe_1480 from pediococcus pentosaceus atcc 25745
|
| 17 | d1o51a_
|
|
 |
96.6 |
21 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:DUF190/COG1993 |
| 18 | c2dclB_
|
|
 |
95.3 |
26 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical upf0166 protein ph1503;
PDBTitle: structure of ph1503 protein from pyrococcus horikoshii ot3
|
| 19 | c2gx8B_
|
|
 |
92.6 |
25 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:nif3-related protein;
PDBTitle: the crystal stucture of bacillus cereus protein related to nif3
|
| 20 | d2gx8a1
|
|
 |
90.7 |
25 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
| 21 | c2nydB_ |
|
not modelled |
86.6 |
19 |
PDB header:unknown function
Chain: B: PDB Molecule:upf0135 protein sa1388;
PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
|
| 22 | d1nh8a2 |
|
not modelled |
84.3 |
9 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain |
| 23 | c3hluA_ |
|
not modelled |
77.5 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein duf2179;
PDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 duf2179 from eubacterium ventriosum
|
| 24 | c2vd3B_ |
|
not modelled |
75.3 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: the structure of histidine inhibited hisg from2 methanobacterium thermoautotrophicum
|
| 25 | c1nh7A_ |
|
not modelled |
71.7 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: atp phosphoribosyltransferase (atp-prtase) from mycobacterium2 tuberculosis
|
| 26 | d1h3da2 |
|
not modelled |
60.3 |
13 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain |
| 27 | c3dfeA_ |
|
not modelled |
59.8 |
25 |
PDB header:signaling protein
Chain: A: PDB Molecule:putative pii-like signaling protein;
PDBTitle: crystal structure of a putative pii-like signaling protein2 (yp_323533.1) from anabaena variabilis atcc 29413 at 2.35 a3 resolution
|
| 28 | c3rrkA_ |
|
not modelled |
47.1 |
16 |
PDB header:proton transport
Chain: A: PDB Molecule:v-type atpase 116 kda subunit;
PDBTitle: crystal structure of the cytoplasmic n-terminal domain of subunit i,2 homolog of subunit a, of v-atpase
|
| 29 | c1q1kA_ |
|
not modelled |
44.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: structure of atp-phosphoribosyltransferase from e. coli complexed with2 pr-atp
|
| 30 | c2f06B_ |
|
not modelled |
32.5 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
|
| 31 | c2cz9A_ |
|
not modelled |
26.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:probable galactokinase;
PDBTitle: crystal structure of galactokinase from pyrococcus horikoshi
|
| 32 | d2hmfa2 |
|
not modelled |
24.9 |
20 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 33 | d1w1oa1 |
|
not modelled |
24.4 |
16 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Cytokinin dehydrogenase 1 |
| 34 | c2zvyB_ |
|
not modelled |
23.5 |
12 |
PDB header:membrane protein
Chain: B: PDB Molecule:chemotaxis protein motb;
PDBTitle: structure of the periplasmic domain of motb from salmonella2 (crystal form ii)
|
| 35 | c2hk3A_ |
|
not modelled |
22.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:diphosphomevalonate decarboxylase;
PDBTitle: crystal structure of mevalonate diphosphate decarboxylase2 from staphylococcus aureus (orthorhombic form)
|
| 36 | d1zhva2 |
|
not modelled |
21.2 |
19 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Atu0741-like |
| 37 | d2j0wa3 |
|
not modelled |
19.3 |
4 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 38 | c2dtjA_ |
|
not modelled |
19.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of regulatory subunit of aspartate kinase2 from corynebacterium glutamicum
|
| 39 | c3mahA_ |
|
not modelled |
19.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:aspartokinase;
PDBTitle: a putative c-terminal regulatory domain of aspartate kinase from2 porphyromonas gingivalis w83.
|
| 40 | d2f06a2 |
|
not modelled |
17.4 |
28 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 41 | d1wjwa_ |
|
not modelled |
17.2 |
30 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
| 42 | c2nuxB_ |
|
not modelled |
15.4 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconate
PDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
|
| 43 | d1v6za2 |
|
not modelled |
15.3 |
23 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YggJ C-terminal domain-like |
| 44 | d2cdqa3 |
|
not modelled |
15.3 |
24 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 45 | d1x8da1 |
|
not modelled |
14.3 |
17 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:YiiL-like |
| 46 | c3fluD_ |
|
not modelled |
13.4 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 47 | c2qlxA_ |
|
not modelled |
13.3 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:l-rhamnose mutarotase;
PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum in complex with l-rhamnose
|
| 48 | c2qlwA_ |
|
not modelled |
13.3 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:rhau;
PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum
|
| 49 | c1zr6A_ |
|
not modelled |
12.4 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucooligosaccharide oxidase;
PDBTitle: the crystal structure of an acremonium strictum glucooligosaccharide2 oxidase reveals a novel flavinylation
|
| 50 | c3dz1A_ |
|
not modelled |
12.4 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
| 51 | c3nuhB_ |
|
not modelled |
12.1 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:dna gyrase subunit b;
PDBTitle: a domain insertion in e. coli gyrb adopts a novel fold that plays a2 critical role in gyrase function
|
| 52 | c3n2xB_ |
|
not modelled |
11.8 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
| 53 | d1vi2a2 |
|
not modelled |
11.8 |
17 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 54 | d2a5la1 |
|
not modelled |
11.0 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 55 | c3nnqA_ |
|
not modelled |
11.0 |
12 |
PDB header:viral protein
Chain: A: PDB Molecule:n-terminal domain of moloney murine leukemia virus
PDBTitle: crystal structure of the n-terminal domain of moloney murine leukemia2 virus integrase, northeast structural genomics consortium target or3
|
| 56 | c2hfvA_ |
|
not modelled |
10.7 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein rpa1041;
PDBTitle: solution nmr structure of protein rpa1041 from pseudomonas2 aeruginosa. northeast structural genomics consortium3 target pat90.
|
| 57 | c2yxgD_ |
|
not modelled |
10.3 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
|
| 58 | c2re1A_ |
|
not modelled |
10.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:aspartokinase, alpha and beta subunits;
PDBTitle: crystal structure of aspartokinase alpha and beta subunits
|
| 59 | d1v7ra_ |
|
not modelled |
10.0 |
32 |
Fold:Anticodon-binding domain-like
Superfamily:ITPase-like
Family:ITPase (Ham1) |
| 60 | c3lerA_ |
|
not modelled |
10.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 61 | d1mwza_ |
|
not modelled |
9.4 |
19 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 62 | c2y3rC_ |
|
not modelled |
9.3 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:taml;
PDBTitle: structure of the tirandamycin-bound fad-dependent2 tirandamycin oxidase taml in p21 space group
|
| 63 | d1p77a2 |
|
not modelled |
9.3 |
21 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 64 | d1fx0a2 |
|
not modelled |
9.2 |
25 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:N-terminal domain of alpha and beta subunits of F1 ATP synthase
Family:N-terminal domain of alpha and beta subunits of F1 ATP synthase |
| 65 | d2i0ka2 |
|
not modelled |
9.0 |
13 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 66 | d1p5dx4 |
|
not modelled |
9.0 |
23 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
| 67 | d1nvta2 |
|
not modelled |
9.0 |
33 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 68 | c3pueA_ |
|
not modelled |
8.8 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 69 | d1nxza2 |
|
not modelled |
8.8 |
20 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YggJ C-terminal domain-like |
| 70 | c2zodB_ |
|
not modelled |
8.5 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase;
PDBTitle: crystal structure of selenophosphate synthetase from2 aquifex aeolicus
|
| 71 | c2gs8A_ |
|
not modelled |
8.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:mevalonate pyrophosphate decarboxylase;
PDBTitle: structure of mevalonate pyrophosphate decarboxylase from streptococcus2 pyogenes
|
| 72 | c1wveB_ |
|
not modelled |
8.4 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-cresol dehydrogenase [hydroxylating]
PDBTitle: p-cresol methylhydroxylase: alteration of the structure of2 the flavoprotein subunit upon its binding to the3 cytochrome subunit
|
| 73 | d1ny5a2 |
|
not modelled |
8.3 |
24 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 74 | c1f0xA_ |
|
not modelled |
8.3 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: crystal structure of d-lactate dehydrogenase, a peripheral2 membrane respiratory enzyme.
|
| 75 | c2ipiD_ |
|
not modelled |
8.3 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aclacinomycin oxidoreductase (aknox);
PDBTitle: crystal structure of aclacinomycin oxidoreductase
|
| 76 | d1nyta2 |
|
not modelled |
8.3 |
25 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 77 | c3b4uB_ |
|
not modelled |
8.3 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
|
| 78 | c3lciA_ |
|
not modelled |
8.1 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
| 79 | d1sqsa_ |
|
not modelled |
8.0 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Hypothetical protein SP1951 |
| 80 | c2bvfA_ |
|
not modelled |
8.0 |
22 |
PDB header:oxidase
Chain: A: PDB Molecule:6-hydroxy-d-nicotine oxidase;
PDBTitle: crystal structure of 6-hydoxy-d-nicotine oxidase from2 arthrobacter nicotinovorans. crystal form 3 (p1)
|
| 81 | c2otnB_ |
|
not modelled |
8.0 |
29 |
PDB header:isomerase
Chain: B: PDB Molecule:diaminopimelate epimerase;
PDBTitle: crystal structure of the catalytically active form of diaminopimelate2 epimerase from bacillus anthracis
|
| 82 | c1z85B_ |
|
not modelled |
7.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:hypothetical protein tm1380;
PDBTitle: crystal structure of a predicted rna methyltransferase (tm1380) from2 thermotoga maritima msb8 at 2.12 a resolution
|
| 83 | c3pm9A_ |
|
not modelled |
7.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative dehydrogenase (rpa1076) from2 rhodopseudomonas palustris cga009 at 2.57 a resolution
|
| 84 | c3cprB_ |
|
not modelled |
7.6 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
| 85 | c3daqB_ |
|
not modelled |
7.6 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
|
| 86 | c3popD_ |
|
not modelled |
7.5 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gilr oxidase;
PDBTitle: the crystal structure of gilr, an oxidoreductase that catalyzes the2 terminal step of gilvocarcin biosynthesis
|
| 87 | c2hfuB_ |
|
not modelled |
7.5 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:mevalonate kinase, putative;
PDBTitle: crystal structure of l. major mevalonate kinase in complex2 with r-mevalonate
|
| 88 | d1vhka2 |
|
not modelled |
7.3 |
17 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YggJ C-terminal domain-like |
| 89 | c2zhoB_ |
|
not modelled |
7.2 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of the regulatory subunit of aspartate2 kinase from thermus thermophilus (ligand free form)
|
| 90 | c2cx8B_ |
|
not modelled |
7.2 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:methyl transferase;
PDBTitle: crystal structure of methyltransferase with ligand(sah)
|
| 91 | c2wdwB_ |
|
not modelled |
7.1 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative hexose oxidase;
PDBTitle: the native crystal structure of the primary hexose oxidase (2 dbv29) in antibiotic a40926 biosynthesis
|
| 92 | c1zhvA_ |
|
not modelled |
7.1 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein atu0741;
PDBTitle: x-ray crystal structure protein atu0741 from agobacterium tumefaciens.2 northeast structural genomics consortium target atr8.
|
| 93 | c3oonA_ |
|
not modelled |
7.0 |
20 |
PDB header:membrane protein
Chain: A: PDB Molecule:outer membrane protein (tpn50);
PDBTitle: the structure of an outer membrance protein from borrelia burgdorferi2 b31
|
| 94 | c3fwaA_ |
|
not modelled |
7.0 |
21 |
PDB header:flavoprotein
Chain: A: PDB Molecule:reticuline oxidase;
PDBTitle: structure of berberine bridge enzyme, c166a variant in complex with2 (s)-reticuline
|
| 95 | d1uxya1 |
|
not modelled |
6.9 |
18 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase (MurB), N-terminal domain |
| 96 | d1xxxa1 |
|
not modelled |
6.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 97 | c2apnA_ |
|
not modelled |
6.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein hi1723;
PDBTitle: hi1723 solution structure
|
| 98 | c1i19B_ |
|
not modelled |
6.7 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cholesterol oxidase;
PDBTitle: crystal structure of cholesterol oxidase from b.sterolicum
|
| 99 | d1e8ga2 |
|
not modelled |
6.5 |
18 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |