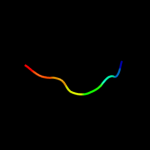
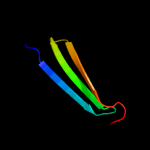
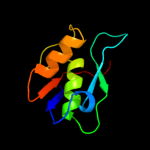
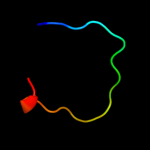
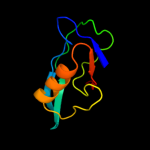
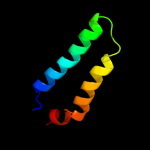
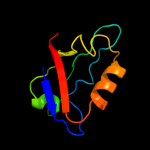
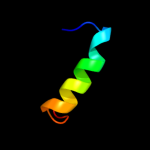
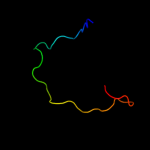
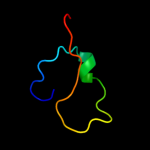
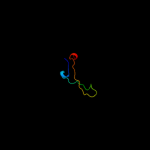
1 d2hrva_
47.0
75
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold2 c1z8rA_
43.3
75
PDB header: hydrolaseChain: A: PDB Molecule: coxsackievirus b4 polyprotein;PDBTitle: 2a cysteine proteinase from human coxsackievirus b4 (strain2 jvb / benschoten / new york / 51)
3 d1bupa2
34.2
31
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP704 d1jcea2
23.3
28
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP705 c2he4A_
23.3
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: na(+)/h(+) exchange regulatory cofactor nhe-rf2;PDBTitle: the crystal structure of the second pdz domain of human2 nherf-2 (slc9a3r2) interacting with a mode 1 pdz binding3 motif
6 c3dgsA_
21.5
32
PDB header: viral proteinChain: A: PDB Molecule: coat protein a;PDBTitle: changing the determinants of protein stability from2 covalent to non-covalent interactions by in-vitro3 evolution: a structural and energetic analysis
7 c3nvbA_
21.0
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of n-terminal part of the protein bf1531 from2 bacteroides fragilis containing phosphatase domain complexed with mg3 and tungstate
8 d2hf3a1
16.8
33
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP709 d1bxla_
16.1
22
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death10 c2v90E_
15.6
15
PDB header: protein-bindingChain: E: PDB Molecule: pdz domain-containing protein 3;PDBTitle: crystal structure of the 3rd pdz domain of intestine- and2 kidney-enriched pdz domain ikepp (pdzd3)
11 d1ysga1
14.6
22
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death12 d1be9a_
14.3
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain13 d2c4ka1
14.3
11
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like14 c2jr3A_
13.6
55
PDB header: antimicrobial proteinChain: A: PDB Molecule: pelovaterin;PDBTitle: antibacterial peptide from eggshell matrix: structure and2 self-assembly of beta-defensin like peptide from the3 chinese soft-shelled turtle eggshell
15 d1t95a3
13.6
21
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: Hypothetical protein AF0491, C-terminal domain16 d2naca2
13.3
19
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain17 d1p9qc3
12.9
21
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: Hypothetical protein AF0491, C-terminal domain18 d2e8aa2
12.9
36
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP7019 c2vyaB_
12.8
35
PDB header: hydrolaseChain: B: PDB Molecule: fatty-acid amide hydrolase 1;PDBTitle: crystal structure of fatty acid amide hydrolase conjugated2 with the drug-like inhibitor pf-750
20 c1g0dA_
12.2
39
PDB header: transferaseChain: A: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: crystal structure of red sea bream transglutaminase
21 d1q59a_
not modelled
11.4
20
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death22 d1dkua1
not modelled
11.3
12
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like23 d1x6ma_
not modelled
11.1
44
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: Glutathione-dependent formaldehyde-activating enzyme, Gfa24 c2d90A_
not modelled
10.9
22
PDB header: protein bindingChain: A: PDB Molecule: pdz domain containing protein 1;PDBTitle: solution structure of the third pdz domain of pdz domain2 containing protein 1
25 c1dkrB_
not modelled
10.8
13
PDB header: transferaseChain: B: PDB Molecule: phosphoribosyl pyrophosphate synthetase;PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
26 d1pbya2
not modelled
10.5
29
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 227 c1l9mB_
not modelled
10.5
32
PDB header: transferaseChain: B: PDB Molecule: protein-glutamine glutamyltransferase e3;PDBTitle: three-dimensional structure of the human transglutaminase 32 enzyme: binding of calcium ions change structure for3 activation
28 c3g9bA_
not modelled
10.5
11
PDB header: transferaseChain: A: PDB Molecule: dolichyl-diphosphooligosaccharide-proteinPDBTitle: crystal structure of reduced ost6l
29 c3pk1A_
not modelled
10.5
11
PDB header: apoptosis/apoptosis regulatorChain: A: PDB Molecule: induced myeloid leukemia cell differentiation protein mcl-PDBTitle: crystal structure of mcl-1 in complex with the baxbh3 domain
30 d3eeqa2
not modelled
10.1
11
Fold: CbiG N-terminal domain-likeSuperfamily: CbiG N-terminal domain-likeFamily: CbiG N-terminal domain-like31 d1g5ma_
not modelled
9.9
20
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death32 d1zy3a1
not modelled
9.9
22
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death33 d2ponb1
not modelled
9.9
22
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death34 c2k2wA_
not modelled
9.5
46
PDB header: cell cycleChain: A: PDB Molecule: recombination and dna repair protein;PDBTitle: second brct domain of nbs1
35 c2dazA_
not modelled
9.5
12
PDB header: protein bindingChain: A: PDB Molecule: inad-like protein;PDBTitle: solution structure of the 7th pdz domain of inad-like2 protein
36 d1wmaa1
not modelled
9.4
29
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases37 c2yv6A_
not modelled
9.4
22
PDB header: apoptosisChain: A: PDB Molecule: bcl-2 homologous antagonist/killer;PDBTitle: crystal structure of human bcl-2 family protein bak
38 d1o0la_
not modelled
9.1
24
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death39 d1f16a_
not modelled
9.0
13
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death40 d1q5xa_
not modelled
9.0
40
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like41 c3qbrA_
not modelled
8.8
13
PDB header: apoptosisChain: A: PDB Molecule: sjchgc06286 protein;PDBTitle: bakbh3 in complex with sja
42 d1ln4a_
not modelled
8.8
29
Fold: IF3-likeSuperfamily: YhbY-likeFamily: YhbY-like43 d1ul1x1
not modelled
8.6
11
Fold: SAM domain-likeSuperfamily: 5' to 3' exonuclease, C-terminal subdomainFamily: 5' to 3' exonuclease, C-terminal subdomain44 d1n7ea_
not modelled
8.5
14
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain45 c3q87A_
not modelled
8.3
67
PDB header: transferase activator/transferaseChain: A: PDB Molecule: putative uncharacterized protein ecu08_1170;PDBTitle: structure of eukaryotic translation termination complex2 methyltransferase mtq2-trm112
46 d1l0wa2
not modelled
8.1
15
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain47 d2i7pa2
not modelled
8.1
8
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Fumble-like48 d1k8ka1
not modelled
8.0
33
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP7049 d1q18a1
not modelled
8.0
25
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Glucokinase50 d1jmxa2
not modelled
8.0
25
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 251 d1l8wa_
not modelled
7.7
8
Fold: Variable surface antigen VlsESuperfamily: Variable surface antigen VlsEFamily: Variable surface antigen VlsE52 c1l8wA_
not modelled
7.7
8
PDB header: immune systemChain: A: PDB Molecule: vlse1;PDBTitle: crystal structure of lyme disease variable surface antigen2 vlse of borrelia burgdorferi
53 c2nacA_
not modelled
7.6
19
PDB header: oxidoreductase(aldehyde(d),nad+(a))Chain: A: PDB Molecule: nad-dependent formate dehydrogenase;PDBTitle: high resolution structures of holo and apo formate dehydrogenase
54 d1roda_
not modelled
7.6
24
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines55 d1mt5a_
not modelled
7.4
41
Fold: Amidase signature (AS) enzymesSuperfamily: Amidase signature (AS) enzymesFamily: Amidase signature (AS) enzymes56 c2egkC_
not modelled
7.4
13
PDB header: protein bindingChain: C: PDB Molecule: general receptor for phosphoinositides 1-PDBTitle: crystal structure of tamalin pdz-intrinsic ligand fusion2 protein
57 d1dkgd2
not modelled
7.3
56
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP7058 d1c0aa2
not modelled
7.3
19
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain59 c2ph7B_
not modelled
7.2
33
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein af_2093;PDBTitle: crystal structure of af2093 from archaeoglobus fulgidus
60 d1u9ya1
not modelled
7.2
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like61 c3hfwA_
not modelled
7.1
27
PDB header: hydrolaseChain: A: PDB Molecule: protein adp-ribosylarginine hydrolase;PDBTitle: crystal structure of human adp-ribosylhydrolase 1 (harh1)
62 d1v4va_
not modelled
7.1
21
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDP-N-acetylglucosamine 2-epimerase63 d1vpra1
not modelled
6.8
75
Fold: LipocalinsSuperfamily: LipocalinsFamily: Dinoflagellate luciferase repeat64 c2xa0A_
not modelled
6.8
19
PDB header: apoptosisChain: A: PDB Molecule: apoptosis regulator bcl-2;PDBTitle: crystal structure of bcl-2 in complex with a bax bh32 peptide
65 c2nrhA_
not modelled
6.5
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: transcriptional activator, putative, baf family;PDBTitle: crystal structure of conserved putative baf family2 transcriptional activator from campylobacter jejuni
66 c2jkzB_
not modelled
6.5
17
PDB header: transferaseChain: B: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: saccharomyces cerevisiae hypoxanthine-guanine2 phosphoribosyltransferase in complex with gmp (guanosine 5'3 -monophosphate) (orthorhombic crystal form)
67 c3eeqB_
not modelled
6.3
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative cobalamin biosynthesis protein gPDBTitle: crystal structure of a putative cobalamin biosynthesis2 protein g homolog from sulfolobus solfataricus
68 c3c8oB_
not modelled
6.1
47
PDB header: hydrolase regulatorChain: B: PDB Molecule: regulator of ribonuclease activity a;PDBTitle: the crystal structure of rraa from pao1
69 d1bwva2
not modelled
6.0
22
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase70 c2o2fA_
not modelled
6.0
20
PDB header: apoptosisChain: A: PDB Molecule: apoptosis regulator bcl-2;PDBTitle: solution structure of the anti-apoptotic protein bcl-2 in2 complex with an acyl-sulfonamide-based ligand
71 c3lnoA_
not modelled
6.0
33
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of domain of unknown function duf59 from2 bacillus anthracis
72 c1kv3F_
not modelled
5.9
50
PDB header: transferaseChain: F: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: human tissue transglutaminase in gdp bound form
73 c2yuyA_
not modelled
5.8
14
PDB header: signaling proteinChain: A: PDB Molecule: rho gtpase activating protein 21;PDBTitle: solution structure of pdz domain of rho gtpase activating2 protein 21
74 c3iraA_
not modelled
5.5
46
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein;PDBTitle: the crystal structure of one domain of the conserved protein from2 methanosarcina mazei go1
75 c2dc0A_
not modelled
5.4
34
PDB header: hydrolaseChain: A: PDB Molecule: probable amidase;PDBTitle: crystal structure of amidase
76 d1rxwa1
not modelled
5.4
14
Fold: SAM domain-likeSuperfamily: 5' to 3' exonuclease, C-terminal subdomainFamily: 5' to 3' exonuclease, C-terminal subdomain77 d1j9ba_
not modelled
5.4
13
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: ArsC-like78 d8ruca2
not modelled
5.3
7
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase79 d1ee8a1
not modelled
5.3
33
Fold: S13-like H2TH domainSuperfamily: S13-like H2TH domainFamily: Middle domain of MutM-like DNA repair proteins80 d1ujva_
not modelled
5.3
36
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain81 d1svda2
not modelled
5.1
13
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase82 c2gx5B_
not modelled
5.1
5
PDB header: transcriptionChain: B: PDB Molecule: gtp-sensing transcriptional pleiotropic repressor cody;PDBTitle: n-terminal gaf domain of transcriptional pleiotropic repressor cody
83 d1vi4a_
not modelled
5.1
27
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like84 d1pq1a_
not modelled
5.0
20
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death85 d2jm6b1
not modelled
5.0
13
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death86 d1wjpa2
not modelled
5.0
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2