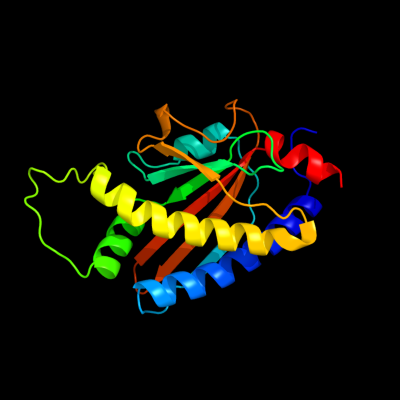
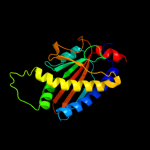
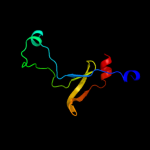
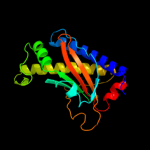
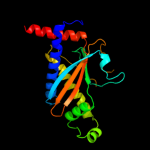
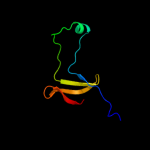
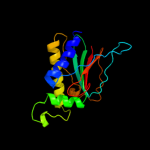
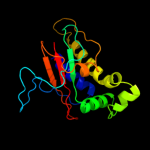
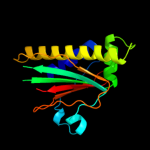
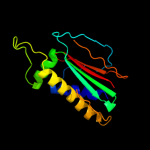
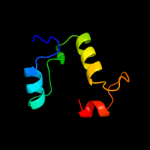
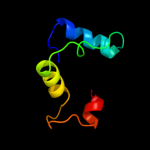
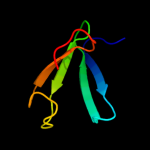
1 c2hp7A_
100.0
26
PDB header: signaling proteinChain: A: PDB Molecule: flagellar motor switch protein flim;PDBTitle: structure of flim provides insight into assembly of the2 switch complex in the bacterial flagella motor
2 d1o6aa_
99.7
18
Fold: Surface presentation of antigens (SPOA)Superfamily: Surface presentation of antigens (SPOA)Family: Surface presentation of antigens (SPOA)3 d1xkra_
99.6
12
Fold: CheC-likeSuperfamily: CheC-likeFamily: CheC-like4 c3qtaA_
99.6
10
PDB header: attractantChain: A: PDB Molecule: chemotaxis protein chec;PDBTitle: crystal structure of a hypothetical chec-like protein (rrnac0528) from2 haloarcula marismortui atcc 43049 at 2.00 a resolution
5 d1o9ya_
99.6
19
Fold: Surface presentation of antigens (SPOA)Superfamily: Surface presentation of antigens (SPOA)Family: Surface presentation of antigens (SPOA)6 d1xkoa_
97.7
11
Fold: CheC-likeSuperfamily: CheC-likeFamily: CheC-like7 c3hzhB_
97.6
11
PDB header: signaling proteinChain: B: PDB Molecule: chemotaxis operon protein (chex);PDBTitle: crystal structure of the chex-chey-bef3-mg+2 complex from2 borrelia burgdorferi
8 c3hm4A_
97.5
14
PDB header: hydrolaseChain: A: PDB Molecule: chemotaxis protein chex;PDBTitle: crystal structure of a chemotaxis protein chex (dde_0281) from2 desulfovibrio desulfuricans subsp. at 1.30 a resolution
9 c3h2dA_
96.1
14
PDB header: signaling proteinChain: A: PDB Molecule: chec-like superfamily protein;PDBTitle: crystal structure of a chemotactic chec-like protein (so_3915) from2 shewanella oneidensis mr-1 at 1.86 a resolution
10 d1bu3a_
58.0
15
Fold: EF Hand-likeSuperfamily: EF-handFamily: Parvalbumin11 c2w85A_
46.1
32
PDB header: protein transportChain: A: PDB Molecule: peroxisomal membrane anchor protein pex14;PDBTitle: structure of pex14 in compex with pex19
12 c3fs7D_
39.3
21
PDB header: metal binding proteinChain: D: PDB Molecule: parvalbumin, thymic;PDBTitle: crystal structure of gallus gallus beta-parvalbumin (avian2 thymic hormone)
13 d2evra2
37.0
16
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: NlpC/P6014 c2pl9D_
28.3
33
PDB header: signaling protienChain: D: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of chey-mg(2+)-bef(3)(-) in complex with chez(c19)2 peptide solved from a p2(1)2(1)2 crystal
15 c2pl9E_
27.9
38
PDB header: signaling protienChain: E: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of chey-mg(2+)-bef(3)(-) in complex with chez(c19)2 peptide solved from a p2(1)2(1)2 crystal
16 c2pl9F_
27.9
38
PDB header: signaling protienChain: F: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of chey-mg(2+)-bef(3)(-) in complex with chez(c19)2 peptide solved from a p2(1)2(1)2 crystal
17 c2fg0B_
24.0
16
PDB header: hydrolaseChain: B: PDB Molecule: cog0791: cell wall-associated hydrolases (invasion-PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
18 d2isba1
23.7
48
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: FumA C-terminal domain-likeFamily: FumA C-terminal domain-like19 c2jijA_
18.3
29
PDB header: hydrolaseChain: A: PDB Molecule: prolyl-4 hydroxylase;PDBTitle: crystal structure of the apo form of chlamydomonas2 reinhardtii prolyl-4 hydroxylase type i
20 c2kycA_
17.2
25
PDB header: calcium binding proteinChain: A: PDB Molecule: parvalbumin, thymic cpv3;PDBTitle: solution structure of ca-free chicken parvalbumin 3 (cpv3)
21 c3jstA_
not modelled
14.8
20
PDB header: lyaseChain: A: PDB Molecule: putative pterin-4-alpha-carbinolamine dehydratase;PDBTitle: crystal structure of transcriptional coactivator/pterin dehydratase2 from brucella melitensis
22 d1rwya_
not modelled
14.5
15
Fold: EF Hand-likeSuperfamily: EF-handFamily: Parvalbumin23 d1a0pa2
not modelled
13.9
33
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Lambda integrase-like, catalytic core24 c3ff5B_
not modelled
13.4
36
PDB header: protein transportChain: B: PDB Molecule: peroxisomal biogenesis factor 14;PDBTitle: crystal structure of the conserved n-terminal domain of the2 peroxisomal matrix-protein-import receptor, pex14p
25 c2vywA_
not modelled
13.2
38
PDB header: oxygen bindingChain: A: PDB Molecule: hemoglobin;PDBTitle: hemoglobin (hb2) from trematode fasciola hepatica
26 c2e63A_
not modelled
13.0
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: kiaa1787 protein;PDBTitle: solution structure of the neuz domain in kiaa1787 protein
27 d1vyua1
not modelled
12.6
21
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain28 d1pvaa_
not modelled
11.9
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Parvalbumin29 c2f9jP_
not modelled
11.8
50
PDB header: rna binding proteinChain: P: PDB Molecule: splicing factor 3b subunit 1;PDBTitle: 3.0 angstrom resolution structure of a y22m mutant of the spliceosomal2 protein p14 bound to a region of sf3b155
30 d1d1na_
not modelled
11.8
25
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors31 c3nkhB_
not modelled
11.0
18
PDB header: recombinationChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of integrase from mrsa strain staphylococcus aureus
32 c2glwA_
not modelled
10.6
27
PDB header: transcriptionChain: A: PDB Molecule: 92aa long hypothetical protein;PDBTitle: the solution structure of phs018 from pyrococcus horikoshii
33 d2cqaa1
not modelled
10.5
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TIP49 domain34 c3peiA_
not modelled
9.9
21
PDB header: hydrolaseChain: A: PDB Molecule: cytosol aminopeptidase;PDBTitle: crystal structure of cytosol aminopeptidase from francisella2 tularensis
35 d1wpga1
not modelled
9.7
24
Fold: Double-stranded beta-helixSuperfamily: Calcium ATPase, transduction domain AFamily: Calcium ATPase, transduction domain A36 d5pala_
not modelled
9.6
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Parvalbumin37 d1khda1
not modelled
9.4
18
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain38 c2uzhB_
not modelled
9.0
25
PDB header: lyaseChain: B: PDB Molecule: 2c-methyl-d-erythritol 2,4-cyclodiphosphatePDBTitle: mycobacterium smegmatis 2c-methyl-d-erythritol-2,4-2 cyclodiphosphate synthase (ispf)
39 d1jz8a3
not modelled
8.8
14
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain40 d1s04a_
not modelled
8.7
21
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain41 c3fkhB_
not modelled
8.5
29
PDB header: oxidoreductaseChain: B: PDB Molecule: putative pyridoxamine 5'-phosphate oxidase;PDBTitle: crystal structure of putative pyridoxamine 5'-phosphate oxidase2 (np_601736.1) from corynebacterium glutamicum atcc 13032 kitasato at3 2.51 a resolution
42 c2pmzV_
not modelled
8.5
10
PDB header: translation, transferaseChain: V: PDB Molecule: dna-directed rna polymerase subunit h;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
43 d2elca1
not modelled
8.2
13
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain44 c3q4nA_
not modelled
8.1
30
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein mj0754;PDBTitle: crystal structure of hypothetical protein mj0754 from methanococcus2 jannaschii dsm 2661
45 c2kx2A_
not modelled
8.0
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the solution structure of mth1821
46 d1nr9a_
not modelled
8.0
33
Fold: FAHSuperfamily: FAHFamily: FAH47 c1i7oC_
not modelled
7.9
35
PDB header: isomerase, lyaseChain: C: PDB Molecule: 4-hydroxyphenylacetate degradation bifunctionalPDBTitle: crystal structure of hpce
48 c3ouiA_
not modelled
7.8
11
PDB header: oxidoreductaseChain: A: PDB Molecule: egl nine homolog 1;PDBTitle: phd2-r717 with 40787422
49 c2krgA_
not modelled
7.4
36
PDB header: signaling proteinChain: A: PDB Molecule: na(+)/h(+) exchange regulatory cofactor nhe-rf1;PDBTitle: solution structure of human sodium/ hydrogen exchange2 regulatory factor 1(150-358)
50 c2dfuB_
not modelled
7.2
32
PDB header: isomeraseChain: B: PDB Molecule: probable 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;PDBTitle: crystal structure of the 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from thermus thermophilus hb8
51 d1eika_
not modelled
7.0
10
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB552 c2ebbA_
not modelled
7.0
33
PDB header: lyaseChain: A: PDB Molecule: pterin-4-alpha-carbinolamine dehydratase;PDBTitle: crystal structure of pterin-4-alpha-carbinolamine2 dehydratase (pterin carbinolamine dehydratase) from3 geobacillus kaustophilus hta426
53 c3itqB_
not modelled
7.0
36
PDB header: oxidoreductaseChain: B: PDB Molecule: prolyl 4-hydroxylase, alpha subunit domain protein;PDBTitle: crystal structure of a prolyl 4-hydroxylase from bacillus anthracis
54 c2hc8A_
not modelled
6.7
17
PDB header: transport proteinChain: A: PDB Molecule: cation-transporting atpase, p-type;PDBTitle: structure of the a. fulgidus copa a-domain
55 c3cp3A_
not modelled
6.7
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of conserved protein of unknown function dip18742 from corynebacterium diphtheriae
56 d1jjcb4
not modelled
6.7
14
Fold: Ferredoxin-likeSuperfamily: Anticodon-binding domain of PheRSFamily: Anticodon-binding domain of PheRS57 d1dzfa2
not modelled
6.6
13
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB558 c2oxgE_
not modelled
6.6
20
PDB header: transport proteinChain: E: PDB Molecule: soxz protein;PDBTitle: the soxyz complex of paracoccus pantotrophus
59 d1hmja_
not modelled
6.5
19
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB560 c3rfuC_
not modelled
6.4
13
PDB header: hydrolase, membrane proteinChain: C: PDB Molecule: copper efflux atpase;PDBTitle: crystal structure of a copper-transporting pib-type atpase
61 d1u5la_
not modelled
6.4
19
Fold: Prion-likeSuperfamily: Prion-likeFamily: Prion-like62 d1b10a_
not modelled
6.1
25
Fold: Prion-likeSuperfamily: Prion-likeFamily: Prion-like63 d1yq2a3
not modelled
6.0
16
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain64 d1v8ga1
not modelled
5.9
14
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain65 d1v8ha1
not modelled
5.8
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: SoxZ-like66 d1tqba_
not modelled
5.8
25
Fold: Prion-likeSuperfamily: Prion-likeFamily: Prion-like67 d1dcpa_
not modelled
5.8
19
Fold: DCoH-likeSuperfamily: PCD-likeFamily: PCD-like68 d1uw3a_
not modelled
5.7
25
Fold: Prion-likeSuperfamily: Prion-likeFamily: Prion-like69 d1u69a_
not modelled
5.7
40
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: 3-demethylubiquinone-9 3-methyltransferase70 d1xyua_
not modelled
5.7
25
Fold: Prion-likeSuperfamily: Prion-likeFamily: Prion-like71 d1xyka_
not modelled
5.5
25
Fold: Prion-likeSuperfamily: Prion-likeFamily: Prion-like72 d1sawa_
not modelled
5.5
32
Fold: FAHSuperfamily: FAHFamily: FAH73 c2zjtB_
not modelled
5.4
21
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit b;PDBTitle: crystal structure of dna gyrase b' domain sheds lights on2 the mechanism for t-segment navigation
74 d2vbua1
not modelled
5.3
32
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin kinase-likeFamily: CTP-dependent riboflavin kinase-like75 d1vgga_
not modelled
5.3
17
Fold: Ta1353-likeSuperfamily: Ta1353-likeFamily: Ta1353-like76 d1ibxa_
not modelled
5.2
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain77 c2gl0A_
not modelled
5.1
21
PDB header: transferaseChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: structure of pae2307 in complex with adenosine
78 d1b8la_
not modelled
5.1
26
Fold: EF Hand-likeSuperfamily: EF-handFamily: Parvalbumin79 d1uoua1
not modelled
5.1
17
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain80 d1ru0a_
not modelled
5.0
15
Fold: DCoH-likeSuperfamily: PCD-likeFamily: PCD-like81 d1xnea_
not modelled
5.0
29
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain82 d2tpta1
not modelled
5.0
25
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain