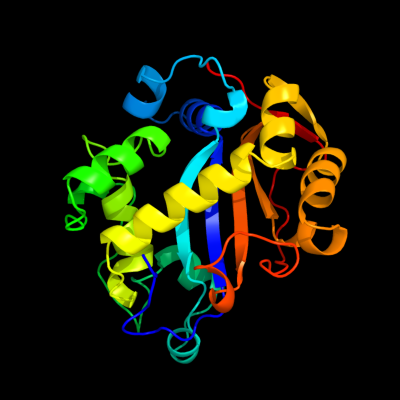
| 1 | c2pqxA_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease i;
PDBTitle: e. coli rnase 1 (in vivo folded)
|
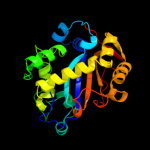
| 2 | d1sgla_
|
|
 |
100.0 |
20 |
Fold:Ribonuclease Rh-like
Superfamily:Ribonuclease Rh-like
Family:Ribonuclease Rh-like |
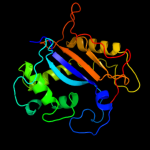
| 3 | d1iyba_
|
|
 |
100.0 |
23 |
Fold:Ribonuclease Rh-like
Superfamily:Ribonuclease Rh-like
Family:Ribonuclease Rh-like |
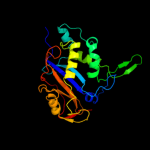
| 4 | d1dixa_
|
|
 |
100.0 |
19 |
Fold:Ribonuclease Rh-like
Superfamily:Ribonuclease Rh-like
Family:Ribonuclease Rh-like |
| 5 | c1vd3A_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:rnase ngr3;
PDBTitle: ribonuclease nt in complex with 2'-ump
|
| 6 | d1jy5a_
|
|
 |
100.0 |
21 |
Fold:Ribonuclease Rh-like
Superfamily:Ribonuclease Rh-like
Family:Ribonuclease Rh-like |
| 7 | d1iooa_
|
|
 |
100.0 |
24 |
Fold:Ribonuclease Rh-like
Superfamily:Ribonuclease Rh-like
Family:Ribonuclease Rh-like |
| 8 | d1ucda_
|
|
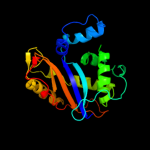 |
100.0 |
24 |
Fold:Ribonuclease Rh-like
Superfamily:Ribonuclease Rh-like
Family:Ribonuclease Rh-like |
| 9 | d1iqqa_
|
|
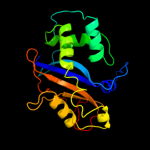 |
100.0 |
20 |
Fold:Ribonuclease Rh-like
Superfamily:Ribonuclease Rh-like
Family:Ribonuclease Rh-like |
| 10 | c3d3zA_
|
|
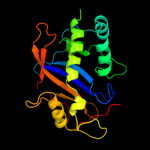 |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:actibind;
PDBTitle: crystal structure of actibind a t2 rnase
|
| 11 | d1bola_
|
|
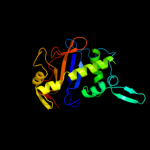 |
100.0 |
19 |
Fold:Ribonuclease Rh-like
Superfamily:Ribonuclease Rh-like
Family:Ribonuclease Rh-like |
| 12 | d2ciwa1
|
|
 |
26.8 |
26 |
Fold:EF Hand-like
Superfamily:Cloroperoxidase
Family:Cloroperoxidase |
| 13 | d1dfxa1
|
|
 |
26.8 |
50 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Superoxide reductase-like
Family:Superoxide reductase-like |
| 14 | c2amuA_
|
|
 |
25.3 |
80 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative superoxide reductase;
PDBTitle: crystal structure of a putative superoxide reductase (tm0658) from2 thermotoga maritima at 2.00 a resolution
|
| 15 | d1dqia_
|
|
 |
24.9 |
80 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Superoxide reductase-like
Family:Superoxide reductase-like |
| 16 | c1y07A_
|
|
 |
23.7 |
50 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:desulfoferrodoxin (rbo);
PDBTitle: crystal structure of the superoxide reductase from2 treponema pallidum
|
| 17 | d1vzia1
|
|
 |
23.7 |
60 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Superoxide reductase-like
Family:Superoxide reductase-like |
| 18 | c1vzhB_
|
|
 |
20.2 |
60 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:desulfoferrodoxin;
PDBTitle: structure of superoxide reductase bound to ferrocyanide and2 active site expansion upon x-ray induced photoreduction
|
| 19 | c3oqgA_
|
|
 |
14.4 |
38 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:hpy188i;
PDBTitle: restriction endonuclease hpy188i in complex with substrate dna
|
| 20 | c2cj0A_
|
|
 |
14.1 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chloroperoxidase;
PDBTitle: chloroperoxidase complexed with nitrate
|
| 21 | c2zkrs_ |
|
not modelled |
12.8 |
14 |
PDB header:ribosomal protein/rna
Chain: S: PDB Molecule:rna expansion segment es39 part ii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 22 | c1s1iT_ |
|
not modelled |
10.3 |
11 |
PDB header:ribosome
Chain: T: PDB Molecule:60s ribosomal protein l25;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
|
| 23 | c2cfaB_ |
|
not modelled |
9.3 |
7 |
PDB header:transferase
Chain: B: PDB Molecule:thymidylate synthase;
PDBTitle: structure of viral flavin-dependant thymidylate synthase2 thyx
|
| 24 | c3bboV_ |
|
not modelled |
9.2 |
11 |
PDB header:ribosome
Chain: V: PDB Molecule:ribosomal protein l23;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
|
| 25 | d2dl2a1 |
|
not modelled |
8.8 |
10 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 26 | c4a10A_ |
|
not modelled |
8.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:octenoyl-coa reductase/carboxylase;
PDBTitle: apo-structure of 2-octenoyl-coa carboxylase reductase cinf from2 streptomyces sp.
|
| 27 | d1m4ka1 |
|
not modelled |
8.3 |
10 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 28 | c2j42A_ |
|
not modelled |
8.2 |
23 |
PDB header:toxin
Chain: A: PDB Molecule:c2 toxin component-ii;
PDBTitle: low quality crystal structure of the transport component c2-2 ii of the c2-toxin from clostridium botulinum
|
| 29 | c4a17R_ |
|
not modelled |
7.8 |
15 |
PDB header:ribosome
Chain: R: PDB Molecule:rpl23a;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
|
| 30 | c3q13A_ |
|
not modelled |
7.7 |
22 |
PDB header:cell adhesion
Chain: A: PDB Molecule:spondin-1;
PDBTitle: the structure of the ca2+-binding, glycosylated f-spondin domain of f-2 spondin, a c2-domain variant from extracellular matrix
|
| 31 | d1nkra1 |
|
not modelled |
7.2 |
10 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 32 | d1h7na_ |
|
not modelled |
6.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 33 | c3f9xA_ |
|
not modelled |
6.7 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:histone-lysine n-methyltransferase setd8;
PDBTitle: structural insights into lysine multiple methylation by set2 domain methyltransferases, set8-y334f / h4-lys20me2 /3 adohcy
|
| 34 | d1vqos1 |
|
not modelled |
6.2 |
11 |
Fold:Ribosomal proteins S24e, L23 and L15e
Superfamily:Ribosomal proteins S24e, L23 and L15e
Family:L23p |
| 35 | c1xa0B_ |
|
not modelled |
5.8 |
7 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative nadph dependent oxidoreductases;
PDBTitle: crystal structure of mcsg target apc35536 from bacillus2 stearothermophilus
|
| 36 | d1f66c_ |
|
not modelled |
5.6 |
16 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 37 | c1f66C_ |
|
not modelled |
5.6 |
16 |
PDB header:structural protein/dna
Chain: C: PDB Molecule:histone h2a.z;
PDBTitle: 2.6 a crystal structure of a nucleosome core particle2 containing the variant histone h2a.z
|
| 38 | d2qamt1 |
|
not modelled |
5.5 |
9 |
Fold:Ribosomal proteins S24e, L23 and L15e
Superfamily:Ribosomal proteins S24e, L23 and L15e
Family:L23p |
| 39 | d1tp6a_ |
|
not modelled |
5.5 |
9 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:PA1314-like |
| 40 | d1xtpa_ |
|
not modelled |
5.2 |
9 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:AD-003 protein-like |
| 41 | d1m4ua_ |
|
not modelled |
5.2 |
56 |
Fold:Cystine-knot cytokines
Superfamily:Cystine-knot cytokines
Family:Noggin |
| 42 | c2ww9K_ |
|
not modelled |
5.1 |
13 |
PDB header:ribosome
Chain: K: PDB Molecule:60s ribosomal protein l25;
PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
|