| 1 |
|
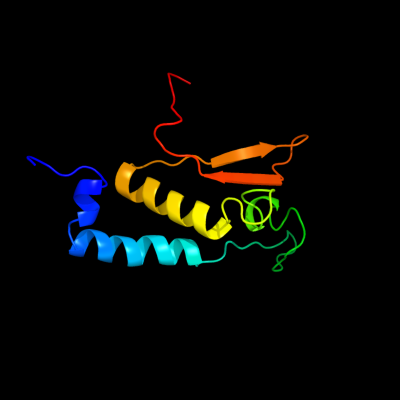
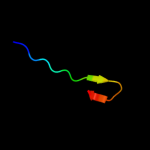
PDB 3kdr chain C
Region: 34 - 140
Aligned: 97
Modelled: 107
Confidence: 92.1%
Identity: 13%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hk97 family phage portal protein;
PDBTitle: the crystal structure of a hk97 family phage portal protein from2 corynebacterium diphtheriae to 2.9a
Phyre2
| 2 |
|
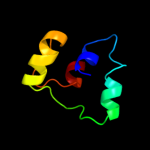
PDB 3fo8 chain D
Region: 114 - 148
Aligned: 35
Modelled: 35
Confidence: 16.1%
Identity: 26%
PDB header:viral protein
Chain: D: PDB Molecule:tail sheath protein gp18;
PDBTitle: crystal structure of the bacteriophage t4 tail sheath2 protein, protease resistant fragment gp18pr
Phyre2
| 3 |
|
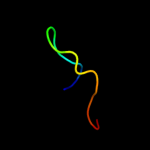
PDB 1c0m chain A domain 1
Region: 105 - 131
Aligned: 27
Modelled: 27
Confidence: 8.1%
Identity: 15%
Fold: SH3-like barrel
Superfamily: DNA-binding domain of retroviral integrase
Family: DNA-binding domain of retroviral integrase
Phyre2
| 4 |
|
PDB 1hq0 chain A
Region: 111 - 125
Aligned: 15
Modelled: 15
Confidence: 6.4%
Identity: 27%
Fold: CNF1/YfiH-like putative cysteine hydrolases
Superfamily: CNF1/YfiH-like putative cysteine hydrolases
Family: Type 1 cytotoxic necrotizing factor, catalytic domain
Phyre2
| 5 |
|
PDB 3zrh chain A
Region: 50 - 104
Aligned: 52
Modelled: 55
Confidence: 6.0%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:ubiquitin thioesterase zranb1;
PDBTitle: crystal structure of the lys29, lys33-linkage-specific trabid otu2 deubiquitinase domain reveals an ankyrin-repeat ubiquitin binding3 domain (ankubd)
Phyre2
| 6 |
|
PDB 3hjd chain A
Region: 141 - 157
Aligned: 17
Modelled: 17
Confidence: 5.1%
Identity: 41%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:human neutrophil peptide 1;
PDBTitle: x-ray structure of monomeric variant of hnp1
Phyre2
| 7 |
|
PDB 3hjd chain B
Region: 141 - 157
Aligned: 17
Modelled: 17
Confidence: 5.1%
Identity: 41%
PDB header:antimicrobial protein
Chain: B: PDB Molecule:human neutrophil peptide 1;
PDBTitle: x-ray structure of monomeric variant of hnp1
Phyre2