| 1 |
|
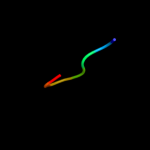
PDB 1d1h chain A
Region: 29 - 35
Aligned: 7
Modelled: 7
Confidence: 12.7%
Identity: 57%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: omega toxin-like
Family: Spider toxins
Phyre2
| 2 |
|
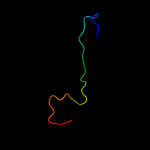
PDB 2i5y chain R
Region: 5 - 33
Aligned: 29
Modelled: 29
Confidence: 12.7%
Identity: 17%
PDB header:virus/viral protein/immune system
Chain: R: PDB Molecule:antibody 17b heavy chain;
PDBTitle: crystal structure of cd4m47, a scorpion-toxin mimic of cd4, in complex2 with hiv-1 yu2 gp120 envelope glycoprotein and anti-hiv-1 antibody3 17b
Phyre2
| 3 |
|
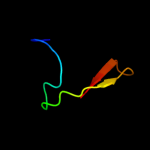
PDB 1mqt chain A
Region: 9 - 37
Aligned: 29
Modelled: 29
Confidence: 12.4%
Identity: 38%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: Positive stranded ssRNA viruses
Family: Picornaviridae-like VP (VP1, VP2, VP3 and VP4)
Phyre2
| 4 |
|
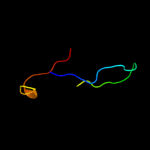
PDB 1pft chain A
Region: 4 - 30
Aligned: 27
Modelled: 27
Confidence: 10.9%
Identity: 15%
Fold: Rubredoxin-like
Superfamily: Zinc beta-ribbon
Family: Transcriptional factor domain
Phyre2
| 5 |
|
PDB 1cz8 chain H
Region: 3 - 33
Aligned: 31
Modelled: 31
Confidence: 10.9%
Identity: 23%
PDB header:immune system
Chain: H: PDB Molecule:heavy chain of neutralizing antibody;
PDBTitle: vascular endothelial growth factor in complex with an2 affinity matured antibody
Phyre2
| 6 |
|
PDB 1xbw chain A
Region: 1 - 14
Aligned: 14
Modelled: 14
Confidence: 8.7%
Identity: 36%
Fold: Ferredoxin-like
Superfamily: Dimeric alpha+beta barrel
Family: PG130-like
Phyre2
| 7 |
|
PDB 2aq2 chain A domain 1
Region: 7 - 33
Aligned: 27
Modelled: 27
Confidence: 7.4%
Identity: 11%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: V set domains (antibody variable domain-like)
Phyre2
| 8 |
|
PDB 2o1u chain A
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 6.9%
Identity: 40%
PDB header:chaperone
Chain: A: PDB Molecule:endoplasmin;
PDBTitle: structure of full length grp94 with amp-pnp bound
Phyre2
| 9 |
|
PDB 2a1h chain A domain 1
Region: 23 - 50
Aligned: 22
Modelled: 28
Confidence: 6.9%
Identity: 23%
Fold: D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily: D-aminoacid aminotransferase-like PLP-dependent enzymes
Family: D-aminoacid aminotransferase-like PLP-dependent enzymes
Phyre2
| 10 |
|
PDB 2xk0 chain A
Region: 28 - 35
Aligned: 8
Modelled: 8
Confidence: 6.3%
Identity: 38%
PDB header:transcription
Chain: A: PDB Molecule:polycomb protein pcl;
PDBTitle: solution structure of the tudor domain from drosophila2 polycomblike (pcl)
Phyre2
| 11 |
|
PDB 2qqr chain A domain 2
Region: 27 - 35
Aligned: 9
Modelled: 9
Confidence: 5.8%
Identity: 56%
Fold: SH3-like barrel
Superfamily: Tudor/PWWP/MBT
Family: Tudor domain
Phyre2
| 12 |
|
PDB 2qqs chain A domain 2
Region: 27 - 35
Aligned: 9
Modelled: 9
Confidence: 5.7%
Identity: 56%
Fold: SH3-like barrel
Superfamily: Tudor/PWWP/MBT
Family: Tudor domain
Phyre2
| 13 |
|
PDB 1uh6 chain A
Region: 13 - 20
Aligned: 8
Modelled: 8
Confidence: 5.7%
Identity: 50%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Ubiquitin-like
Family: Ubiquitin-related
Phyre2
| 14 |
|
PDB 1tiu chain A
Region: 7 - 35
Aligned: 29
Modelled: 29
Confidence: 5.5%
Identity: 21%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: I set domains
Phyre2
| 15 |
|
PDB 2hug chain A domain 1
Region: 30 - 35
Aligned: 6
Modelled: 6
Confidence: 5.4%
Identity: 67%
Fold: SH3-like barrel
Superfamily: Chromo domain-like
Family: Chromo domain
Phyre2
| 16 |
|
PDB 2oqj chain B
Region: 6 - 33
Aligned: 28
Modelled: 28
Confidence: 5.2%
Identity: 25%
PDB header:immune system
Chain: B: PDB Molecule:fab 2g12 heavy chain;
PDBTitle: crystal structure analysis of fab 2g12 in complex with peptide 2g12.1
Phyre2