| 1 | c2b664_ |
|
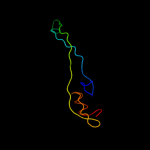 |
100.0 |
32 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: 50s ribosomal subunit from a crystal structure of release factor rf1,2 trnas and mrna bound to the ribosome. this file contains the 50s3 subunit from a crystal structure of release factor rf1, trnas and4 mrna bound to the ribosome and is described in remark 400
|
| 2 | d1vs6z1 |
|
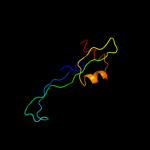 |
100.0 |
46 |
Fold:L28p-like
Superfamily:L28p-like
Family:Ribosomal protein L31p |
| 3 | c3bbo1_ |
|
 |
100.0 |
28 |
PDB header:ribosome
Chain: 1: PDB Molecule:ribosomal protein l31;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
|
| 4 | d2j0141 |
|
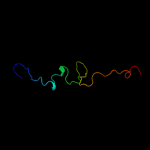 |
99.9 |
39 |
Fold:L28p-like
Superfamily:L28p-like
Family:Ribosomal protein L31p |
| 5 | c2j034_ |
|
 |
99.9 |
39 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 4 of 4).3 this file contains the 50s subunit from molecule ii.
|
| 6 | c3f1f4_ |
|
 |
97.1 |
27 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: crystal structure of a translation termination complex2 formed with release factor rf2. this file contains the 50s3 subunit of one 70s ribosome. the entire crystal structure4 contains two 70s ribosomes as described in remark 400.
|
| 7 | c2wh44_ |
|
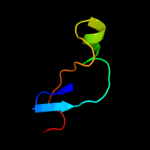 |
97.1 |
27 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: insights into translational termination from the structure2 of rf2 bound to the ribosome
|
| 8 | c2ketA_ |
|
 |
29.0 |
31 |
PDB header:antibiotic
Chain: A: PDB Molecule:cathelicidin-6;
PDBTitle: solution structure of bmap-27
|
| 9 | d1x6ha1 |
|
 |
27.6 |
16 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 10 | c3cucB_ |
|
 |
21.9 |
54 |
PDB header:signaling protein
Chain: B: PDB Molecule:protein of unknown function with a fic domain;
PDBTitle: crystal structure of a fic domain containing signaling protein2 (bt_2513) from bacteroides thetaiotaomicron vpi-5482 at 2.71 a3 resolution
|
| 11 | d1tqza1 |
|
 |
20.7 |
25 |
Fold:PH domain-like barrel
Superfamily:PH domain-like
Family:Necap1 N-terminal domain-like |
| 12 | c3g66A_ |
|
 |
17.8 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:sortase c;
PDBTitle: the crystal structure of streptococcus pneumoniae sortase c2 provides novel insights into catalysis as well as pilin3 substrate specificity
|
| 13 | c3cngC_ |
|
 |
16.6 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:nudix hydrolase;
PDBTitle: crystal structure of nudix hydrolase from nitrosomonas europaea
|
| 14 | d1eb7a2 |
|
 |
16.5 |
25 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c peroxidase |
| 15 | d2fiya1 |
|
 |
16.5 |
0 |
Fold:FdhE-like
Superfamily:FdhE-like
Family:FdhE-like |
| 16 | c3o0pA_ |
|
 |
15.3 |
11 |
PDB header:transferase , hydrolase
Chain: A: PDB Molecule:sortase family protein;
PDBTitle: pilus-related sortase c of group b streptococcus
|
| 17 | c4a1eT_ |
|
 |
15.2 |
26 |
PDB header:ribosome
Chain: T: PDB Molecule:rpl24;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
|
| 18 | d1nmla2 |
|
 |
15.0 |
17 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c peroxidase |
| 19 | c2yiuE_ |
|
 |
14.7 |
43 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
|
| 20 | c3cwbQ_ |
|
 |
13.6 |
43 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:mitochondrial cytochrome c1, heme protein;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
|
| 21 | c2w1jB_ |
|
not modelled |
13.5 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:putative sortase;
PDBTitle: crystal structure of sortase c-1 (srtc-1) from2 streptococcus pneumoniae
|
| 22 | c3n3vA_ |
|
not modelled |
12.7 |
46 |
PDB header:transferase
Chain: A: PDB Molecule:adenosine monophosphate-protein transferase ibpa;
PDBTitle: crystal structure of ibpafic2-h3717a in complex with adenylylated2 cdc42
|
| 23 | c2fynH_ |
|
not modelled |
12.5 |
29 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:cytochrome c1;
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
|
| 24 | c1nmlA_ |
|
not modelled |
12.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:di-haem cytochrome c peroxidase;
PDBTitle: di-haemic cytochrome c peroxidase from pseudomonas nautica 617, form2 in (ph 4.0)
|
| 25 | d3cx5d1 |
|
not modelled |
12.5 |
29 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Cytochrome bc1 domain |
| 26 | c2kw8A_ |
|
not modelled |
11.8 |
16 |
PDB header:protein binding
Chain: A: PDB Molecule:lpxtg-site transpeptidase family protein;
PDBTitle: solution structure of bacillus anthracis sortase a (srta)2 transpeptidase
|
| 27 | c1p84D_ |
|
not modelled |
11.6 |
29 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
| 28 | d1ppjd1 |
|
not modelled |
11.5 |
43 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Cytochrome bc1 domain |
| 29 | c1zrtD_ |
|
not modelled |
11.2 |
43 |
PDB header:oxidoreductase/metal transport
Chain: D: PDB Molecule:cytochrome c1;
PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
|
| 30 | c3hq7A_ |
|
not modelled |
10.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c551 peroxidase;
PDBTitle: ccpa from g. sulfurreducens, g94k/k97q/r100i variant
|
| 31 | c1dvbA_ |
|
not modelled |
10.7 |
4 |
PDB header:electron transport
Chain: A: PDB Molecule:rubrerythrin;
PDBTitle: rubrerythrin
|
| 32 | c3izcZ_ |
|
not modelled |
10.7 |
19 |
PDB header:ribosome
Chain: Z: PDB Molecule:60s ribosomal protein rpl24 (l24e);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 33 | c2b9vB_ |
|
not modelled |
10.6 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-amino acid ester hydrolase;
PDBTitle: acetobacter turbidans alpha-amino acid ester hydrolase
|
| 34 | d1oa8a_ |
|
not modelled |
10.1 |
20 |
Fold:AXH domain
Superfamily:AXH domain
Family:AXH domain |
| 35 | d1e7la1 |
|
not modelled |
10.1 |
60 |
Fold:LEM/SAP HeH motif
Superfamily:Recombination endonuclease VII, C-terminal and dimerization domains
Family:Recombination endonuclease VII, C-terminal and dimerization domains |
| 36 | c2keqA_ |
|
not modelled |
9.7 |
36 |
PDB header:splicing
Chain: A: PDB Molecule:dna polymerase iii alpha subunit, nucleic acid
PDBTitle: solution structure of dnae intein from nostoc punctiforme
|
| 37 | c3iz5Z_ |
|
not modelled |
9.4 |
22 |
PDB header:ribosome
Chain: Z: PDB Molecule:60s ribosomal protein l24 (l24e);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 38 | d1mi8a_ |
|
not modelled |
9.3 |
25 |
Fold:Hedgehog/intein (Hint) domain
Superfamily:Hedgehog/intein (Hint) domain
Family:Intein (protein splicing domain) |
| 39 | c1zd7B_ |
|
not modelled |
8.7 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:dna polymerase iii alpha subunit;
PDBTitle: 1.7 angstrom crystal structure of post-splicing form of a dnae intein2 from synechocystis sp. pcc 6803
|
| 40 | c3mdnD_ |
|
not modelled |
8.6 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:glutamine aminotransferase class-ii domain protein;
PDBTitle: structure of glutamine aminotransferase class-ii domain protein2 (spo2029) from silicibacter pomeroyi
|
| 41 | c3rbjB_ |
|
not modelled |
8.5 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:sortase family protein;
PDBTitle: crystal structure of the lid-mutant of streptococcus agalactiae2 sortase c1
|
| 42 | d1iufa2 |
|
not modelled |
8.3 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Centromere-binding |
| 43 | c3eqxB_ |
|
not modelled |
8.3 |
46 |
PDB header:dna binding protein
Chain: B: PDB Molecule:fic domain containing transcriptional regulator;
PDBTitle: crystal structure of a fic family protein (so_4266) from shewanella2 oneidensis at 1.6 a resolution
|
| 44 | c1xezA_ |
|
not modelled |
7.8 |
32 |
PDB header:toxin
Chain: A: PDB Molecule:hemolysin;
PDBTitle: crystal structure of the vibrio cholerae cytolysin (hlya)2 pro-toxin with octylglucoside bound
|
| 45 | c2w1kB_ |
|
not modelled |
7.7 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:putative sortase;
PDBTitle: crystal structure of sortase c-3 (srtc-3) from2 streptococcus pneumoniae
|
| 46 | d1wada_ |
|
not modelled |
7.7 |
50 |
Fold:Multiheme cytochromes
Superfamily:Multiheme cytochromes
Family:Cytochrome c3-like |
| 47 | d1at0a_ |
|
not modelled |
7.7 |
21 |
Fold:Hedgehog/intein (Hint) domain
Superfamily:Hedgehog/intein (Hint) domain
Family:Hedgehog C-terminal (Hog) autoprocessing domain |
| 48 | c1iqcB_ |
|
not modelled |
7.3 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:di-heme peroxidase;
PDBTitle: crystal structure of di-heme peroxidase from nitrosomonas europaea
|
| 49 | c1mpxB_ |
|
not modelled |
7.2 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-amino acid ester hydrolase;
PDBTitle: alpha-amino acid ester hydrolase labeled with selenomethionine
|
| 50 | d1am2a_ |
|
not modelled |
7.1 |
20 |
Fold:Hedgehog/intein (Hint) domain
Superfamily:Hedgehog/intein (Hint) domain
Family:Intein (protein splicing domain) |
| 51 | c3dd7A_ |
|
not modelled |
6.8 |
20 |
PDB header:ribosome inhibitor
Chain: A: PDB Molecule:death on curing protein;
PDBTitle: structure of doch66y in complex with the c-terminal domain of phd
|
| 52 | d1j5ya2 |
|
not modelled |
6.6 |
19 |
Fold:HPr-like
Superfamily:Putative transcriptional regulator TM1602, C-terminal domain
Family:Putative transcriptional regulator TM1602, C-terminal domain |
| 53 | c1l7qA_ |
|
not modelled |
6.5 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:cocaine esterase;
PDBTitle: ser117ala mutant of bacterial cocaine esterase coce
|
| 54 | c2lcjA_ |
|
not modelled |
6.5 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:pab polc intein;
PDBTitle: solution nmr structure of pab polii intein
|
| 55 | c2xwgA_ |
|
not modelled |
6.4 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:sortase;
PDBTitle: crystal structure of sortase c-1 from actinomyces oris (formerly2 actinomyces naeslundii)
|
| 56 | c3ougA_ |
|
not modelled |
6.4 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:aspartate 1-decarboxylase;
PDBTitle: crystal structure of cleaved l-aspartate-alpha-decarboxylase from2 francisella tularensis
|
| 57 | c1uheA_ |
|
not modelled |
6.3 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:aspartate 1-decarboxylase alpha chain;
PDBTitle: crystal structure of aspartate decarboxylase, isoaspargine complex
|
| 58 | c2jspA_ |
|
not modelled |
6.2 |
10 |
PDB header:gene regulation
Chain: A: PDB Molecule:transcriptional regulatory protein ros;
PDBTitle: the prokaryotic cys2his2 zinc finger adopts a novel fold as2 revealed by the nmr structure of a. tumefaciens ros dna3 binding domain
|
| 59 | c3re9A_ |
|
not modelled |
6.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:sortase-like protein;
PDBTitle: crystal structure of sortasec1 from streptococcus suis
|
| 60 | c2imzA_ |
|
not modelled |
6.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease pi-mtui;
PDBTitle: crystal structure of mtu reca intein splicing domain
|
| 61 | c3floD_ |
|
not modelled |
5.9 |
15 |
PDB header:transferase
Chain: D: PDB Molecule:dna polymerase alpha catalytic subunit a;
PDBTitle: crystal structure of the carboxyl-terminal domain of yeast2 dna polymerase alpha in complex with its b subunit
|
| 62 | c1iufA_ |
|
not modelled |
5.8 |
25 |
PDB header:dna binding protein
Chain: A: PDB Molecule:centromere abp1 protein;
PDBTitle: low resolution solution structure of the two dna-binding2 domains in schizosaccharomyces pombe abp1 protein
|
| 63 | c2in0A_ |
|
not modelled |
5.8 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease pi-mtui;
PDBTitle: crystal structure of mtu reca intein splicing domain
|
| 64 | c2c45F_ |
|
not modelled |
5.7 |
13 |
PDB header:lyase
Chain: F: PDB Molecule:aspartate 1-decarboxylase precursor;
PDBTitle: native precursor of pyruvoyl dependent aspartate2 decarboxylase
|
| 65 | d1i77a_ |
|
not modelled |
5.6 |
67 |
Fold:Multiheme cytochromes
Superfamily:Multiheme cytochromes
Family:Cytochrome c3-like |
| 66 | c3hq2A_ |
|
not modelled |
5.6 |
42 |
PDB header:hydrolase
Chain: A: PDB Molecule:bacillus subtilis m32 carboxypeptidase;
PDBTitle: bsucp crystal structure
|
| 67 | c2jmzA_ |
|
not modelled |
5.5 |
30 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein mj0781;
PDBTitle: solution structure of a klba intein precursor from2 methanococcus jannaschii
|
| 68 | c1v0eB_ |
|
not modelled |
5.4 |
41 |
PDB header:hydrolase
Chain: B: PDB Molecule:endo-alpha-sialidase;
PDBTitle: endosialidase of bacteriophage k1f
|
| 69 | c3gycB_ |
|
not modelled |
5.2 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative glycoside hydrolase;
PDBTitle: crystal structure of putative glycoside hydrolase (yp_001304622.1)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
|
| 70 | c4znfA_ |
|
not modelled |
5.2 |
21 |
PDB header:zinc finger dna binding domain
Chain: A: PDB Molecule:zinc finger;
PDBTitle: high-resolution three-dimensional structure of a single2 zinc finger from a human enhancer binding protein in3 solution
|
| 71 | c3znfA_ |
|
not modelled |
5.2 |
21 |
PDB header:zinc finger dna binding domain
Chain: A: PDB Molecule:zinc finger;
PDBTitle: high-resolution three-dimensional structure of a single2 zinc finger from a human enhancer binding protein in3 solution
|
| 72 | d4znfa_ |
|
not modelled |
5.2 |
21 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |

