| 1 |
|
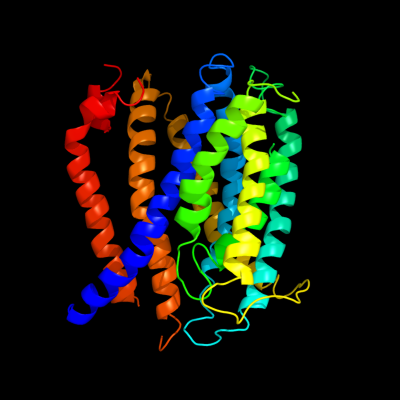
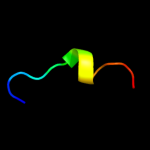
PDB 3mku chain A
Region: 4 - 355
Aligned: 347
Modelled: 347
Confidence: 100.0%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:multi antimicrobial extrusion protein (na(+)/drug
PDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
Phyre2
| 2 |
|
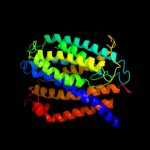
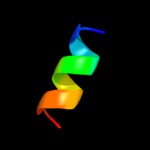
PDB 1mqs chain B
Region: 1 - 7
Aligned: 7
Modelled: 7
Confidence: 22.6%
Identity: 86%
PDB header:endocytosis/exocytosis
Chain: B: PDB Molecule:integral membrane protein sed5;
PDBTitle: crystal structure of sly1p in complex with an n-terminal2 peptide of sed5p
Phyre2
| 3 |
|
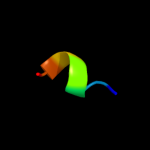
PDB 3j01 chain A
Region: 284 - 336
Aligned: 53
Modelled: 53
Confidence: 8.1%
Identity: 6%
PDB header:ribosome/ribosomal protein
Chain: A: PDB Molecule:preprotein translocase secy subunit;
PDBTitle: structure of the ribosome-secye complex in the membrane environment
Phyre2
| 4 |
|
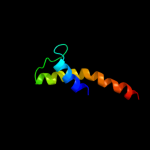
PDB 1jo5 chain A
Region: 305 - 339
Aligned: 35
Modelled: 35
Confidence: 7.4%
Identity: 17%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 5 |
|
PDB 1kdx chain B
Region: 5 - 23
Aligned: 19
Modelled: 19
Confidence: 6.3%
Identity: 11%
PDB header:transcription regulation complex
Chain: B: PDB Molecule:creb;
PDBTitle: kix domain of mouse cbp (creb binding protein) in complex2 with phosphorylated kinase inducible domain (pkid) of rat3 creb (cyclic amp response element binding protein), nmr 174 structures
Phyre2
| 6 |
|
PDB 2vck chain C
Region: 336 - 352
Aligned: 17
Modelled: 17
Confidence: 6.2%
Identity: 24%
PDB header:oxidoreductase
Chain: C: PDB Molecule:cyanobacterial phycoerythrobilin;
PDBTitle: structure of phycoerythrobilin synthase pebs from the2 cyanophage p-ssm2 in complex with the bound substrate3 biliverdin ixa
Phyre2
| 7 |
|
PDB 1u17 chain A domain 1
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 6.0%
Identity: 31%
Fold: Lipocalins
Superfamily: Lipocalins
Family: Retinol binding protein-like
Phyre2
| 8 |
|
PDB 2nzz chain A
Region: 6 - 16
Aligned: 11
Modelled: 11
Confidence: 6.0%
Identity: 36%
PDB header:membrane protein
Chain: A: PDB Molecule:penetratin conjugated gas (374-394) peptide;
PDBTitle: nmr structure analysis of the penetratin conjugated gas2 (374-394) peptide
Phyre2
| 9 |
|
PDB 2r6g chain F domain 2
Region: 5 - 48
Aligned: 44
Modelled: 44
Confidence: 5.9%
Identity: 11%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 10 |
|
PDB 1x8q chain A
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 5.6%
Identity: 31%
Fold: Lipocalins
Superfamily: Lipocalins
Family: Retinol binding protein-like
Phyre2
| 11 |
|
PDB 1akh chain A
Region: 6 - 16
Aligned: 11
Modelled: 11
Confidence: 5.6%
Identity: 27%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Homeodomain
Phyre2