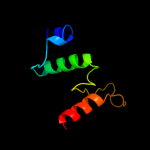
1 c3qe7A_
100.0
17
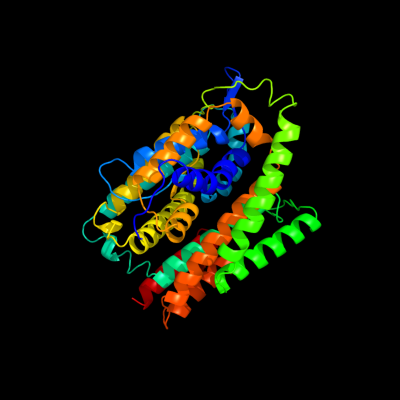
PDB header: transport proteinChain: A: PDB Molecule: uracil permease;PDBTitle: crystal structure of uracil transporter--uraa
2 d2a65a1
29.6
7
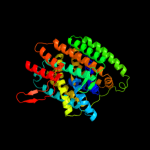
Fold: SNF-likeSuperfamily: SNF-likeFamily: SNF-like3 c3orgB_
23.7
14
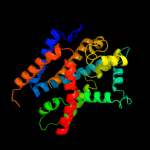
PDB header: transport proteinChain: B: PDB Molecule: cmclc;PDBTitle: crystal structure of a eukaryotic clc transporter
4 c3lpzA_
22.4
0
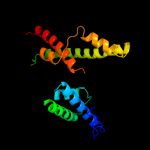
PDB header: protein transportChain: A: PDB Molecule: get4 (yor164c homolog);PDBTitle: crystal structure of c. therm. get4
5 c1unhD_
17.1
23
PDB header: cell cycleChain: D: PDB Molecule: cyclin-dependent kinase 5 activator 1;PDBTitle: structural mechanism for the inhibition of cdk5-p25 by2 roscovitine, aloisine and indirubin.
6 d1unld_
16.4
23
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Cyclin7 c3nd0A_
15.9
16
PDB header: transport proteinChain: A: PDB Molecule: sll0855 protein;PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
8 c1bzkA_
11.6
13
PDB header: transport proteinChain: A: PDB Molecule: protein (band 3 anion transport protein);PDBTitle: structural studies on the effects of the deletion in the2 red cell anion exchanger (band3, ae1) associated with3 south east asian ovalocytosis.
9 c2d4vD_
11.1
12
PDB header: oxidoreductaseChain: D: PDB Molecule: isocitrate dehydrogenase;PDBTitle: crystal structure of nad dependent isocitrate dehydrogenase2 from acidithiobacillus thiooxidans
10 c2bbjB_
10.4
4
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
11 c2kpeA_
9.8
21
PDB header: membrane proteinChain: A: PDB Molecule: glycophorin-a;PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
12 c2kpeB_
9.8
21
PDB header: membrane proteinChain: B: PDB Molecule: glycophorin-a;PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
13 c2ht2B_
9.7
9
PDB header: membrane proteinChain: B: PDB Molecule: h(+)/cl(-) exchange transporter clca;PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
14 c2xteH_
9.1
24
PDB header: transcriptionChain: H: PDB Molecule: f-box-like/wd repeat-containing protein tbl1x;PDBTitle: structure of the tbl1 tetramerisation domain
15 c3fgxA_
8.6
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: rbstp2171;PDBTitle: structure of uncharacterised protein rbstp2171 from bacillus2 stearothermophilus
16 c2xtdB_
8.5
24
PDB header: transcriptionChain: B: PDB Molecule: tbl1 f-box-like/wd repeat-containing protein tbl1x;PDBTitle: structure of the tbl1 tetramerisation domain
17 c1bttA_
8.1
18
PDB header: transmembrane proteinChain: A: PDB Molecule: band 3 anion transport protein;PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
18 c1btsA_
8.1
18
PDB header: transmembrane proteinChain: A: PDB Molecule: band 3 anion transport protein;PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
19 d1v53a1
7.9
7
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases20 d1g2ua_
7.8
7
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases21 c2e0cA_
not modelled
7.4
4
PDB header: oxidoreductaseChain: A: PDB Molecule: 409aa long hypothetical nadp-dependent isocitratePDBTitle: crystal structure of isocitrate dehydrogenase from sulfolobus tokodaii2 strain7 at 2.0 a resolution
22 d1rutx4
not modelled
7.3
38
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain23 c1x0lB_
not modelled
7.2
5
PDB header: oxidoreductaseChain: B: PDB Molecule: homoisocitrate dehydrogenase;PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
24 c3u1hA_
not modelled
7.2
2
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-isopropylmalate dehydrogenase;PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
25 d1xaca_
not modelled
7.1
7
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases26 d1cnza_
not modelled
7.1
5
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases27 c3a0bt_
not modelled
6.3
21
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of br-substituted photosystem ii complex
28 d2axtt1
not modelled
6.3
21
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein T, PsbTFamily: PsbT-like29 c2axtt_
not modelled
6.3
21
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center t protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
30 c3a0ht_
not modelled
6.3
21
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of i-substituted photosystem ii complex
31 c3a0hT_
not modelled
6.3
21
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of i-substituted photosystem ii complex
32 c3kziT_
not modelled
6.3
21
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
33 c2axtT_
not modelled
6.3
21
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center t protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
34 c3arcT_
not modelled
6.3
21
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
35 c1svfB_
not modelled
6.3
31
PDB header: viral proteinChain: B: PDB Molecule: protein (fusion glycoprotein);PDBTitle: paramyxovirus sv5 fusion protein core
36 c1s5lT_
not modelled
6.3
21
PDB header: photosynthesisChain: T: PDB Molecule: photosystem ii psbt protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
37 c1s5lt_
not modelled
6.3
21
PDB header: photosynthesisChain: T: PDB Molecule: photosystem ii psbt protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
38 d1mswd_
not modelled
6.3
44
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: T7 RNA polymerase39 c3prrT_
not modelled
6.3
21
PDB header: photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
40 c3bz1T_
not modelled
6.3
21
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
41 c3prqT_
not modelled
6.3
21
PDB header: photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
42 c3bz2T_
not modelled
6.3
21
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
43 c2qeuA_
not modelled
6.2
7
PDB header: lyaseChain: A: PDB Molecule: putative carboxymuconolactone decarboxylase;PDBTitle: crystal structure of putative carboxymuconolactone decarboxylase2 (yp_555818.1) from burkholderia xenovorans lb400 at 1.65 a resolution
44 c1wazA_
not modelled
6.2
5
PDB header: transport proteinChain: A: PDB Molecule: merf;PDBTitle: nmr structure determination of the bacterial mercury2 transporter, merf, in micelles
45 d1cm7a_
not modelled
6.2
5
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases46 c2pjvA_
not modelled
6.0
22
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: solution structure of hiv-1 gp41 fusion domain bound to dpc2 micelle
47 d1vlca_
not modelled
6.0
2
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases48 d1wpwa_
not modelled
5.9
2
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases49 d2pnga1
not modelled
5.9
10
Fold: Acyl carrier protein-likeSuperfamily: ACP-likeFamily: Acyl-carrier protein (ACP)50 c3s0xB_
not modelled
5.8
13
PDB header: hydrolaseChain: B: PDB Molecule: peptidase a24b, flak domain protein;PDBTitle: the crystal structure of gxgd membrane protease flak
51 c1erfA_
not modelled
5.8
22
PDB header: viral proteinChain: A: PDB Molecule: transmembrane glycoprotein;PDBTitle: conformational mapping of the n-terminal fusion peptide of2 hiv-1 gp41 using 13c-enhanced fourier transform infrared3 spectroscopy (ftir)
52 d1hqsa_
not modelled
5.7
5
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases53 c2gfpA_
not modelled
5.7
14
PDB header: membrane proteinChain: A: PDB Molecule: multidrug resistance protein d;PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
54 c3r8wC_
not modelled
5.6
7
PDB header: oxidoreductaseChain: C: PDB Molecule: 3-isopropylmalate dehydrogenase 2, chloroplastic;PDBTitle: structure of 3-isopropylmalate dehydrogenase isoform 2 from2 arabidopsis thaliana at 2.2 angstrom resolution
55 d1w0da_
not modelled
5.6
10
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases56 c1mxeE_
not modelled
5.5
57
PDB header: metal binding proteinChain: E: PDB Molecule: target sequence of rat calmodulin-dependentPDBTitle: structure of the complex of calmodulin with the target2 sequence of camki
57 c1mxeF_
not modelled
5.5
57
PDB header: metal binding proteinChain: F: PDB Molecule: target sequence of rat calmodulin-dependentPDBTitle: structure of the complex of calmodulin with the target2 sequence of camki
58 c3spaA_
not modelled
5.5
22
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerase, mitochondrial;PDBTitle: crystal structure of human mitochondrial rna polymerase
59 c2jd3B_
not modelled
5.4
7
PDB header: dna binding proteinChain: B: PDB Molecule: stbb protein;PDBTitle: parr from plasmid pb171
60 d1gr0a1
not modelled
5.4
8
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain61 c1afoB_
not modelled
5.4
23
PDB header: integral membrane proteinChain: B: PDB Molecule: glycophorin a;PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
62 d1a05a_
not modelled
5.4
5
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases63 d2vzsa2
not modelled
5.4
38
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain64 d1pb1a_
not modelled
5.4
5
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases65 c3neyC_
not modelled
5.3
23
PDB header: membrane proteinChain: C: PDB Molecule: 55 kda erythrocyte membrane protein;PDBTitle: crystal structure of the kinase domain of mpp1/p55
66 c2qfyE_
not modelled
5.3
2
PDB header: oxidoreductaseChain: E: PDB Molecule: isocitrate dehydrogenase [nadp];PDBTitle: crystal structure of saccharomyces cerevesiae mitochondrial nadp(+)-2 dependent isocitrate dehydrogenase in complex with a-ketoglutarate
67 d3e46a1
not modelled
5.3
40
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain68 d1q90r_
not modelled
5.3
26
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor69 c1q90R_
not modelled
5.3
26
PDB header: photosynthesisChain: R: PDB Molecule: cytochrome b6-f complex iron-sulfur subunit;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
70 c2kncA_
not modelled
5.2
17
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
71 c2d1cB_
not modelled
5.2
7
PDB header: oxidoreductaseChain: B: PDB Molecule: isocitrate dehydrogenase;PDBTitle: crystal structure of tt0538 protein from thermus thermophilus hb8
72 c2kjfA_
not modelled
5.2
23
PDB header: antimicrobial proteinChain: A: PDB Molecule: carnocyclin-a;PDBTitle: the solution structure of the circular bacteriocin2 carnocyclin a (ccla)
73 c2l9xA_
not modelled
5.2
14
PDB header: antimicrobial proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: trn- peptide of the two-component bacteriocin thuricin cd
74 c1b9uA_
not modelled
5.2
14
PDB header: hydrolaseChain: A: PDB Molecule: protein (atp synthase);PDBTitle: membrane domain of the subunit b of the e.coli atp synthase
75 c1btrA_
not modelled
5.2
16
PDB header: anion transportChain: A: PDB Molecule: band 3 anion transport protein;PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
76 c1btqA_
not modelled
5.2
16
PDB header: anion transportChain: A: PDB Molecule: band 3 anion transport protein;PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
77 c3p5nA_
not modelled
5.2
11
PDB header: transport proteinChain: A: PDB Molecule: riboflavin uptake protein;PDBTitle: structure and mechanism of the s component of a bacterial ecf2 transporter