| 1 |
|
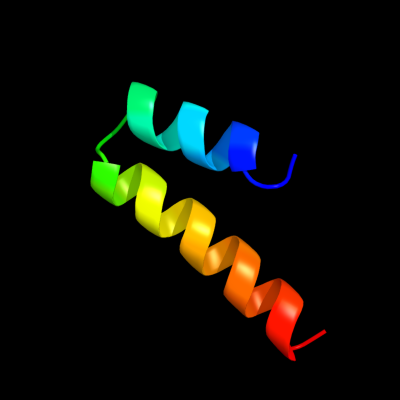
PDB 3p5n chain A
Region: 64 - 95
Aligned: 32
Modelled: 32
Confidence: 66.0%
Identity: 19%
PDB header:transport protein
Chain: A: PDB Molecule:riboflavin uptake protein;
PDBTitle: structure and mechanism of the s component of a bacterial ecf2 transporter
Phyre2
| 2 |
|
PDB 3rko chain M
Region: 11 - 150
Aligned: 137
Modelled: 140
Confidence: 35.2%
Identity: 11%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 3 |
|
PDB 2bbj chain B
Region: 159 - 217
Aligned: 51
Modelled: 59
Confidence: 26.3%
Identity: 14%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 4 |
|
PDB 2iub chain A domain 2
Region: 159 - 218
Aligned: 44
Modelled: 45
Confidence: 14.0%
Identity: 14%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 5 |
|
PDB 3rko chain N
Region: 7 - 156
Aligned: 144
Modelled: 150
Confidence: 9.4%
Identity: 12%
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-quinone oxidoreductase subunit n;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 6 |
|
PDB 1f6g chain A
Region: 1 - 128
Aligned: 124
Modelled: 128
Confidence: 9.2%
Identity: 10%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 7 |
|
PDB 1cii chain A
Region: 116 - 185
Aligned: 65
Modelled: 70
Confidence: 7.0%
Identity: 11%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 8 |
|
PDB 1pw4 chain A
Region: 159 - 224
Aligned: 66
Modelled: 66
Confidence: 6.5%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 9 |
|
PDB 2jln chain A
Region: 31 - 236
Aligned: 193
Modelled: 206
Confidence: 6.4%
Identity: 17%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 10 |
|
PDB 1iij chain A
Region: 157 - 183
Aligned: 27
Modelled: 27
Confidence: 5.9%
Identity: 7%
PDB header:signaling protein
Chain: A: PDB Molecule:erbb-2 receptor protein-tyrosine kinase;
PDBTitle: solution structure of the neu/erbb-2 membrane spanning2 segment
Phyre2
| 11 |
|
PDB 3k3g chain A
Region: 38 - 100
Aligned: 63
Modelled: 63
Confidence: 5.4%
Identity: 8%
PDB header:transport protein
Chain: A: PDB Molecule:urea transporter;
PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
Phyre2
| 12 |
|
PDB 1fft chain B domain 2
Region: 184 - 231
Aligned: 48
Modelled: 48
Confidence: 5.2%
Identity: 8%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 13 |
|
PDB 3dtu chain B domain 2
Region: 181 - 230
Aligned: 50
Modelled: 50
Confidence: 5.2%
Identity: 14%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 14 |
|
PDB 2knc chain A
Region: 184 - 234
Aligned: 51
Modelled: 51
Confidence: 5.0%
Identity: 10%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2