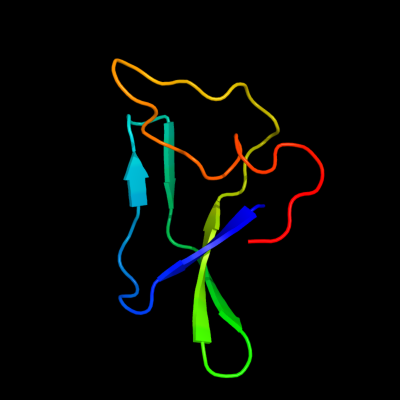
| 1 | c1t3oA_
|
|
 |
99.9 |
41 |
PDB header:rna binding protein
Chain: A: PDB Molecule:carbon storage regulator;
PDBTitle: solution structure of csra, a bacterial carbon storage2 regulatory protein
|
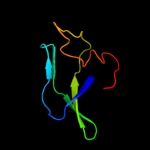
| 2 | c2jppB_
|
|
 |
99.9 |
75 |
PDB header:translation/rna
Chain: B: PDB Molecule:translational repressor;
PDBTitle: structural basis of rsma/csra rna recognition: structure of2 rsme bound to the shine-dalgarno sequence of hcna mrna
|
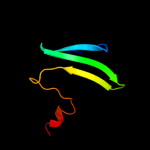
| 3 | d1vpza_
|
|
 |
99.9 |
89 |
Fold:CsrA-like
Superfamily:CsrA-like
Family:CsrA-like |
| 4 | c1vpzB_
|
|
 |
99.9 |
89 |
PDB header:rna binding protein
Chain: B: PDB Molecule:carbon storage regulator homolog;
PDBTitle: crystal structure of a putative carbon storage regulator protein2 (csra, pa0905) from pseudomonas aeruginosa at 2.05 a resolution
|
| 5 | d1e0ta1
|
|
 |
63.3 |
31 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 6 | d2g50a1
|
|
 |
62.7 |
17 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 7 | d1pkma1
|
|
 |
60.6 |
18 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 8 | d1liua1
|
|
 |
59.6 |
21 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 9 | c1e0tD_
|
|
 |
38.5 |
32 |
PDB header:phosphotransferase
Chain: D: PDB Molecule:pyruvate kinase;
PDBTitle: r292d mutant of e. coli pyruvate kinase
|
| 10 | d2o3ga1
|
|
 |
35.6 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 11 | c3eoeC_
|
|
 |
34.2 |
32 |
PDB header:transferase
Chain: C: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pyruvate kinase from toxoplasma gondii, 55.m00007
|
| 12 | d1vhka1
|
|
 |
34.0 |
19 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:YggJ N-terminal domain-like |
| 13 | c3ilaG_
|
|
 |
31.3 |
31 |
PDB header:signaling protein
Chain: G: PDB Molecule:ryanodine receptor 1;
PDBTitle: crystal structure of rabbit ryanodine receptor 1 n-terminal domain (9-2 205)
|
| 14 | c1t5aB_
|
|
 |
30.6 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase, m2 isozyme;
PDBTitle: human pyruvate kinase m2
|
| 15 | d2o1ra1
|
|
 |
30.3 |
29 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 16 | c1aqfB_
|
|
 |
29.3 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase;
PDBTitle: pyruvate kinase from rabbit muscle with mg, k, and l-2 phospholactate
|
| 17 | d1a3xa1
|
|
 |
28.7 |
23 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 18 | c1go3E_
|
|
 |
28.2 |
24 |
PDB header:transferase
Chain: E: PDB Molecule:dna-directed rna polymerase subunit e;
PDBTitle: structure of an archeal homolog of the eukaryotic rna2 polymerase ii rpb4/rpb7 complex
|
| 19 | d1pkla1
|
|
 |
27.8 |
25 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 20 | d1pmia_
|
|
 |
27.3 |
22 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Type I phosphomannose isomerase |
| 21 | c2eqsA_ |
|
not modelled |
26.2 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent rna helicase dhx8;
PDBTitle: solution structure of the s1 rna binding domain of human2 atp-dependent rna helicase dhx8
|
| 22 | c3llbA_ |
|
not modelled |
22.8 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of the protein pa3983 with unknown2 function from pseudomonas aeruginosa pao1
|
| 23 | c3ma8A_ |
|
not modelled |
21.1 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of cgd1_2040, a pyruvate kinase from cryptosporidium2 parvum
|
| 24 | d2r2za1 |
|
not modelled |
20.0 |
16 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 25 | c3ec1A_ |
|
not modelled |
19.6 |
24 |
PDB header:hydrolase, signaling protein
Chain: A: PDB Molecule:yqeh gtpase;
PDBTitle: structure of yqeh gtpase from geobacillus stearothermophilus2 (an atnos1 / atnoa1 ortholog)
|
| 26 | c3dedB_ |
|
not modelled |
19.6 |
13 |
PDB header:membrane protein
Chain: B: PDB Molecule:probable hemolysin;
PDBTitle: c-terminal domain of probable hemolysin from chromobacterium violaceum
|
| 27 | c3h0gS_ |
|
not modelled |
18.3 |
16 |
PDB header:transcription
Chain: S: PDB Molecule:dna-directed rna polymerase ii subunit rpb7;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 28 | d2z0sa1 |
|
not modelled |
18.2 |
18 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 29 | c2xrhA_ |
|
not modelled |
16.5 |
29 |
PDB header:unknown function
Chain: A: PDB Molecule:protein hp0721;
PDBTitle: crystal structure of the truncated form of hp0721
|
| 30 | d3deda1 |
|
not modelled |
15.7 |
13 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 31 | c2khiA_ |
|
not modelled |
14.8 |
24 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s1;
PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
|
| 32 | c1pklB_ |
|
not modelled |
14.6 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:protein (pyruvate kinase);
PDBTitle: the structure of leishmania pyruvate kinase
|
| 33 | d1y14b1 |
|
not modelled |
14.4 |
13 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 34 | d2p13a1 |
|
not modelled |
13.7 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 35 | d2nqwa1 |
|
not modelled |
13.2 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 36 | c2khjA_ |
|
not modelled |
12.9 |
32 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s1;
PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
|
| 37 | d1go3e1 |
|
not modelled |
12.7 |
18 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 38 | d2f23a2 |
|
not modelled |
12.7 |
28 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:GreA transcript cleavage factor, C-terminal domain |
| 39 | c2b8kG_ |
|
not modelled |
12.5 |
13 |
PDB header:transferase
Chain: G: PDB Molecule:dna-directed rna polymerase ii 19 kda
PDBTitle: 12-subunit rna polymerase ii
|
| 40 | d2oaia1 |
|
not modelled |
12.0 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 41 | d1hh2p1 |
|
not modelled |
11.9 |
37 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 42 | c3e0vB_ |
|
not modelled |
11.7 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pyruvate kinase from leishmania mexicana in2 complex with sulphate ions
|
| 43 | d2plsa1 |
|
not modelled |
11.6 |
8 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 44 | d1nxza1 |
|
not modelled |
11.3 |
19 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:YggJ N-terminal domain-like |
| 45 | d1xnea_ |
|
not modelled |
10.9 |
25 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
| 46 | c3h2yA_ |
|
not modelled |
10.3 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:gtpase family protein;
PDBTitle: crystal structure of yqeh gtpase from bacillus anthracis with dgdp2 bound
|
| 47 | d1c0fa1 |
|
not modelled |
10.2 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
| 48 | d2plia1 |
|
not modelled |
10.0 |
25 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 49 | d2c35b1 |
|
not modelled |
9.7 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 50 | c2xoaA_ |
|
not modelled |
9.7 |
31 |
PDB header:metal transport
Chain: A: PDB Molecule:ryanodine receptor 1;
PDBTitle: crystal structure of the n-terminal three domains of the2 skeletal muscle ryanodine receptor (ryr1)
|
| 51 | c3qr5B_ |
|
not modelled |
9.6 |
31 |
PDB header:signaling protein
Chain: B: PDB Molecule:cardiac ca2+ release channel;
PDBTitle: structure of the first domain of a cardiac ryanodine receptor mutant2 with exon 3 deleted
|
| 52 | d2rk5a1 |
|
not modelled |
9.5 |
20 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 53 | d2etna2 |
|
not modelled |
9.4 |
28 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:GreA transcript cleavage factor, C-terminal domain |
| 54 | c2cqoA_ |
|
not modelled |
9.0 |
23 |
PDB header:ribosome
Chain: A: PDB Molecule:nucleolar protein of 40 kda;
PDBTitle: solution structure of the s1 rna binding domain of human2 hypothetical protein flj11067
|
| 55 | c1xzzA_ |
|
not modelled |
8.9 |
38 |
PDB header:membrane protein
Chain: A: PDB Molecule:inositol 1,4,5-trisphosphate receptor type 1;
PDBTitle: crystal structure of the ligand binding suppressor domain of type 12 inositol 1,4,5-trisphosphate receptor
|
| 56 | d2q3za2 |
|
not modelled |
8.2 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
| 57 | c1yz6A_ |
|
not modelled |
8.2 |
20 |
PDB header:translation
Chain: A: PDB Molecule:probable translation initiation factor 2 alpha
PDBTitle: crystal structure of intact alpha subunit of aif2 from2 pyrococcus abyssi
|
| 58 | d1q46a2 |
|
not modelled |
8.1 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 59 | c2z51A_ |
|
not modelled |
7.8 |
14 |
PDB header:metal transport
Chain: A: PDB Molecule:nifu-like protein 2, chloroplast;
PDBTitle: crystal structure of arabidopsis cnfu involved in iron-2 sulfur cluster biosynthesis
|
| 60 | d1hqia_ |
|
not modelled |
7.2 |
22 |
Fold:Monooxygenase (hydroxylase) regulatory protein
Superfamily:Monooxygenase (hydroxylase) regulatory protein
Family:Monooxygenase (hydroxylase) regulatory protein |
| 61 | d1wi5a_ |
|
not modelled |
7.2 |
24 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 62 | d2asba1 |
|
not modelled |
6.6 |
29 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 63 | c2k52A_ |
|
not modelled |
6.0 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mj1198;
PDBTitle: structure of uncharacterized protein mj1198 from2 methanocaldococcus jannaschii. northeast structural3 genomics target mjr117b
|
| 64 | d1kl9a2 |
|
not modelled |
6.0 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 65 | c3t8sA_ |
|
not modelled |
5.7 |
38 |
PDB header:transport protein, membrane protein
Chain: A: PDB Molecule:inositol 1,4,5-trisphosphate receptor type 1;
PDBTitle: apo and insp3-bound crystal structures of the ligand-binding domain of2 an insp3 receptor
|
| 66 | c2c35F_ |
|
not modelled |
5.7 |
16 |
PDB header:polymerase
Chain: F: PDB Molecule:dna-directed rna polymerase ii 19 kda
PDBTitle: subunits rpb4 and rpb7 of human rna polymerase ii
|
| 67 | d2bf5a1 |
|
not modelled |
5.6 |
17 |
Fold:Monooxygenase (hydroxylase) regulatory protein
Superfamily:Monooxygenase (hydroxylase) regulatory protein
Family:Monooxygenase (hydroxylase) regulatory protein |
| 68 | d1gdta1 |
|
not modelled |
5.5 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |
| 69 | d1jmxa3 |
|
not modelled |
5.5 |
30 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Quinohemoprotein amine dehydrogenase A chain, domains 4 and 5 |
| 70 | d2bf2a1 |
|
not modelled |
5.5 |
17 |
Fold:Monooxygenase (hydroxylase) regulatory protein
Superfamily:Monooxygenase (hydroxylase) regulatory protein
Family:Monooxygenase (hydroxylase) regulatory protein |
| 71 | c3l6tB_ |
|
not modelled |
5.3 |
42 |
PDB header:hydrolase
Chain: B: PDB Molecule:mobilization protein trai;
PDBTitle: crystal structure of an n-terminal mutant of the plasmid pcu1 trai2 relaxase domain
|
| 72 | d1ttna1 |
|
not modelled |
5.2 |
14 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 73 | c1ttnA_ |
|
not modelled |
5.2 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:dendritic cell-derived ubiquitin-like protein;
PDBTitle: solution structure of the ubiquitin-like domain of human dc-2 ubp from dendritic cells
|
| 74 | c2mobA_ |
|
not modelled |
5.1 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (methane monooxygenase regulatory
PDBTitle: methane monooxygenase component b
|
| 75 | d2moba_ |
|
not modelled |
5.1 |
11 |
Fold:Monooxygenase (hydroxylase) regulatory protein
Superfamily:Monooxygenase (hydroxylase) regulatory protein
Family:Monooxygenase (hydroxylase) regulatory protein |