| 1 |
|
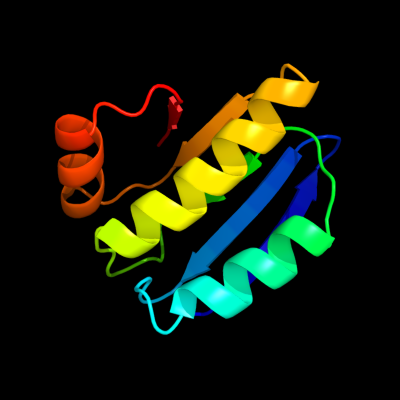
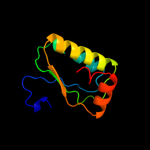
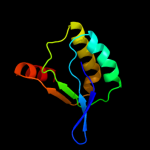
PDB 1th8 chain B
Region: 1 - 96
Aligned: 96
Modelled: 96
Confidence: 99.7%
Identity: 23%
Fold: SpoIIaa-like
Superfamily: SpoIIaa-like
Family: Anti-sigma factor antagonist SpoIIaa
Phyre2
| 2 |
|
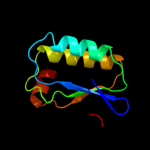
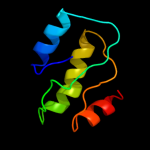
PDB 3t6o chain A
Region: 2 - 96
Aligned: 94
Modelled: 95
Confidence: 99.7%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:sulfate transporter/antisigma-factor antagonist stas;
PDBTitle: the structure of an anti-sigma-factor antagonist (stas) domain protein2 from planctomyces limnophilus.
Phyre2
| 3 |
|
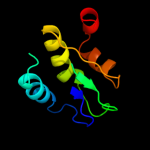
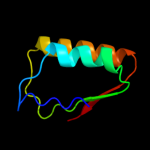
PDB 1vc1 chain A
Region: 1 - 94
Aligned: 94
Modelled: 94
Confidence: 99.7%
Identity: 22%
Fold: SpoIIaa-like
Superfamily: SpoIIaa-like
Family: Anti-sigma factor antagonist SpoIIaa
Phyre2
| 4 |
|
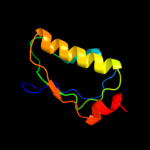
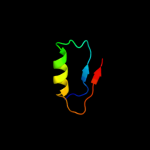
PDB 3f43 chain A
Region: 1 - 96
Aligned: 96
Modelled: 96
Confidence: 99.7%
Identity: 17%
PDB header:transcription
Chain: A: PDB Molecule:putative anti-sigma factor antagonist tm1081;
PDBTitle: crystal structure of a putative anti-sigma factor antagonist (tm1081)2 from thermotoga maritima at 1.59 a resolution
Phyre2
| 5 |
|
PDB 1auz chain A
Region: 1 - 96
Aligned: 96
Modelled: 96
Confidence: 99.7%
Identity: 19%
Fold: SpoIIaa-like
Superfamily: SpoIIaa-like
Family: Anti-sigma factor antagonist SpoIIaa
Phyre2
| 6 |
|
PDB 1h4x chain A
Region: 7 - 94
Aligned: 87
Modelled: 88
Confidence: 99.6%
Identity: 16%
Fold: SpoIIaa-like
Superfamily: SpoIIaa-like
Family: Anti-sigma factor antagonist SpoIIaa
Phyre2
| 7 |
|
PDB 3mgl chain A
Region: 13 - 96
Aligned: 84
Modelled: 84
Confidence: 99.6%
Identity: 20%
PDB header:transport protein
Chain: A: PDB Molecule:sulfate permease family protein;
PDBTitle: crystal structure of permease family protein from vibrio2 cholerae
Phyre2
| 8 |
|
PDB 3oir chain A
Region: 2 - 94
Aligned: 93
Modelled: 93
Confidence: 99.5%
Identity: 18%
PDB header:transport protein
Chain: A: PDB Molecule:sulfate transporter sulfate transporter family protein;
PDBTitle: crystal structure of sulfate transporter family protein from wolinella2 succinogenes
Phyre2
| 9 |
|
PDB 2vy9 chain A
Region: 5 - 91
Aligned: 87
Modelled: 86
Confidence: 99.5%
Identity: 18%
PDB header:gene regulation
Chain: A: PDB Molecule:anti-sigma-factor antagonist;
PDBTitle: molecular architecture of the stressosome, a signal2 integration and transduction hub
Phyre2
| 10 |
|
PDB 2kln chain A
Region: 13 - 96
Aligned: 84
Modelled: 84
Confidence: 99.5%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:probable sulphate-transport transmembrane protein, cog0659;
PDBTitle: solution structure of stas domain of rv1739c from m. tuberculosis
Phyre2
| 11 |
|
PDB 3lkl chain B
Region: 9 - 86
Aligned: 78
Modelled: 78
Confidence: 99.4%
Identity: 19%
PDB header:transport protein
Chain: B: PDB Molecule:antisigma-factor antagonist stas;
PDBTitle: crystal structure of the c-terminal domain of anti-sigma factor2 antagonist stas from rhodobacter sphaeroides
Phyre2
| 12 |
|
PDB 3ny7 chain A
Region: 13 - 96
Aligned: 83
Modelled: 84
Confidence: 99.3%
Identity: 17%
PDB header:membrane protein
Chain: A: PDB Molecule:sulfate transporter;
PDBTitle: stas domain of ychm bound to acp
Phyre2
| 13 |
|
PDB 3llo chain A
Region: 13 - 94
Aligned: 78
Modelled: 82
Confidence: 99.2%
Identity: 14%
PDB header:motor protein
Chain: A: PDB Molecule:prestin;
PDBTitle: crystal structure of the stas domain of motor protein prestin (anion2 transporter slc26a5)
Phyre2
| 14 |
|
PDB 3ih9 chain A
Region: 7 - 79
Aligned: 73
Modelled: 73
Confidence: 95.6%
Identity: 19%
PDB header:hydrolase
Chain: A: PDB Molecule:salt-tolerant glutaminase;
PDBTitle: crystal structure analysis of mglu in its tris form
Phyre2
| 15 |
|
PDB 3c9u chain A domain 2
Region: 50 - 91
Aligned: 41
Modelled: 42
Confidence: 25.7%
Identity: 17%
Fold: PurM C-terminal domain-like
Superfamily: PurM C-terminal domain-like
Family: PurM C-terminal domain-like
Phyre2
| 16 |
|
PDB 2q3l chain A domain 1
Region: 1 - 89
Aligned: 89
Modelled: 89
Confidence: 18.5%
Identity: 18%
Fold: SpoIIaa-like
Superfamily: SpoIIaa-like
Family: Sfri0576-like
Phyre2
| 17 |
|
PDB 3ghf chain A
Region: 19 - 90
Aligned: 67
Modelled: 72
Confidence: 14.0%
Identity: 12%
PDB header:cell cycle
Chain: A: PDB Molecule:septum site-determining protein minc;
PDBTitle: crystal structure of the septum site-determining protein2 minc from salmonella typhimurium
Phyre2
| 18 |
|
PDB 3bez chain C
Region: 13 - 76
Aligned: 64
Modelled: 64
Confidence: 12.1%
Identity: 20%
PDB header:hydrolase
Chain: C: PDB Molecule:protease 4;
PDBTitle: crystal structure of escherichia coli signal peptide peptidase (sppa),2 semet crystals
Phyre2
| 19 |
|
PDB 2fug chain 6 domain 1
Region: 9 - 44
Aligned: 36
Modelled: 36
Confidence: 10.3%
Identity: 14%
Fold: HydA/Nqo6-like
Superfamily: HydA/Nqo6-like
Family: Nq06-like
Phyre2
| 20 |
|
PDB 2pr7 chain A
Region: 40 - 92
Aligned: 53
Modelled: 53
Confidence: 8.3%
Identity: 13%
PDB header:hydrolase
Chain: A: PDB Molecule:haloacid dehalogenase/epoxide hydrolase family;
PDBTitle: crystal structure of uncharacterized protein (np_599989.1) from2 corynebacterium glutamicum atcc 13032 kitasato at 1.44 a resolution
Phyre2
| 21 |
|
| 22 |
|