| 1 |
|
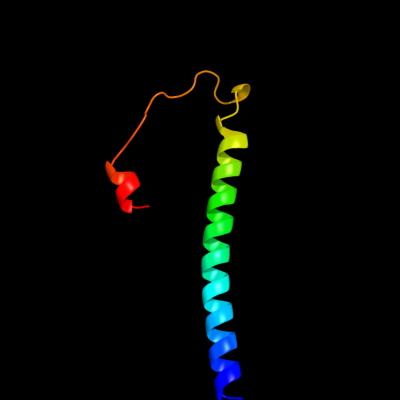
PDB 3ehb chain B domain 2
Region: 163 - 219
Aligned: 57
Modelled: 57
Confidence: 16.9%
Identity: 7%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 2 |
|
PDB 1fft chain B domain 2
Region: 163 - 206
Aligned: 44
Modelled: 44
Confidence: 14.9%
Identity: 14%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 3 |
|
PDB 1ar1 chain B
Region: 163 - 219
Aligned: 57
Modelled: 57
Confidence: 14.3%
Identity: 7%
PDB header:complex (oxidoreductase/antibody)
Chain: B: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
Phyre2
| 4 |
|
PDB 1qle chain B
Region: 163 - 219
Aligned: 57
Modelled: 57
Confidence: 14.3%
Identity: 7%
PDB header:oxidoreductase/immune system
Chain: B: PDB Molecule:cytochrome c oxidase polypeptide ii;
PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
Phyre2
| 5 |
|
PDB 2oso chain A domain 1
Region: 71 - 110
Aligned: 32
Modelled: 37
Confidence: 12.4%
Identity: 16%
Fold: Ligand-binding domain in the NO signalling and Golgi transport
Superfamily: Ligand-binding domain in the NO signalling and Golgi transport
Family: MJ1460-like
Phyre2
| 6 |
|
PDB 2lbg chain A
Region: 62 - 78
Aligned: 17
Modelled: 17
Confidence: 10.9%
Identity: 29%
PDB header:membrane protein
Chain: A: PDB Molecule:major prion protein;
PDBTitle: structure of the chr of the prion protein in dpc micelles
Phyre2
| 7 |
|
PDB 1fft chain G
Region: 163 - 234
Aligned: 72
Modelled: 72
Confidence: 9.6%
Identity: 11%
PDB header:oxidoreductase
Chain: G: PDB Molecule:ubiquinol oxidase;
PDBTitle: the structure of ubiquinol oxidase from escherichia coli
Phyre2
| 8 |
|
PDB 3dtu chain B domain 2
Region: 163 - 219
Aligned: 57
Modelled: 57
Confidence: 9.3%
Identity: 9%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 9 |
|
PDB 2bbj chain B
Region: 95 - 198
Aligned: 70
Modelled: 70
Confidence: 8.8%
Identity: 16%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 10 |
|
PDB 1p58 chain F
Region: 96 - 116
Aligned: 21
Modelled: 21
Confidence: 8.4%
Identity: 24%
PDB header:virus
Chain: F: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
Phyre2
| 11 |
|
PDB 2csb chain A domain 4
Region: 99 - 112
Aligned: 14
Modelled: 14
Confidence: 7.7%
Identity: 14%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Topoisomerase V repeat domain
Phyre2
| 12 |
|
PDB 2l9u chain A
Region: 54 - 80
Aligned: 27
Modelled: 27
Confidence: 6.3%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-3;
PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
Phyre2
| 13 |
|
PDB 1w6k chain A domain 2
Region: 31 - 112
Aligned: 74
Modelled: 82
Confidence: 6.1%
Identity: 9%
Fold: alpha/alpha toroid
Superfamily: Terpenoid cyclases/Protein prenyltransferases
Family: Terpene synthases
Phyre2
| 14 |
|
PDB 1y5i chain C domain 1
Region: 151 - 209
Aligned: 59
Modelled: 59
Confidence: 5.9%
Identity: 14%
Fold: Heme-binding four-helical bundle
Superfamily: Respiratory nitrate reductase 1 gamma chain
Family: Respiratory nitrate reductase 1 gamma chain
Phyre2
| 15 |
|
PDB 2hn8 chain A
Region: 191 - 217
Aligned: 27
Modelled: 27
Confidence: 5.7%
Identity: 26%
PDB header:viral protein
Chain: A: PDB Molecule:protein pb1-f2;
PDBTitle: structural characterization and oligomerization of pb1-f2,2 a pro-apoptotic influenza a virus protein
Phyre2