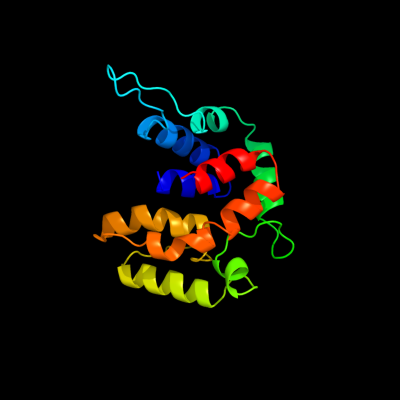
1 d2o9xa1
100.0
24
Fold: TorD-likeSuperfamily: TorD-likeFamily: TorD-like2 c2o9xA_
100.0
22
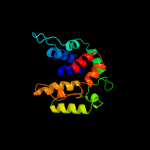
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: reductase, assembly protein;PDBTitle: crystal structure of a putative redox enzyme maturation protein from2 archaeoglobus fulgidus
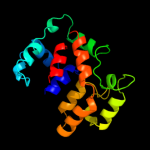
3 d1n1ca_
99.9
22
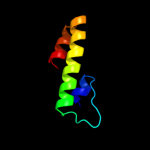
Fold: TorD-likeSuperfamily: TorD-likeFamily: TorD-like4 d1s9ua_
99.9
17
Fold: TorD-likeSuperfamily: TorD-likeFamily: TorD-like5 d2idga1
98.6
23
Fold: TorD-likeSuperfamily: TorD-likeFamily: TorD-like6 d1lvaa3
28.7
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal fragment of elongation factor SelB7 c3ol4B_
17.2
19
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
8 c2di4B_
14.6
16
PDB header: hydrolaseChain: B: PDB Molecule: cell division protein ftsh homolog;PDBTitle: crystal structure of the ftsh protease domain
9 d2ce7a1
11.1
15
Fold: FtsH protease domain-likeSuperfamily: FtsH protease domain-likeFamily: FtsH protease domain-like10 d2ejna2
8.2
22
Fold: Uteroglobin-likeSuperfamily: Uteroglobin-likeFamily: Uteroglobin-like11 d2d6fc2
7.9
27
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain12 c2kvcA_
7.6
25
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
13 d2g3aa1
7.5
28
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT14 d1ghea_
7.3
28
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT15 c3ka1A_
7.1
13
PDB header: chaperoneChain: A: PDB Molecule: rbcx protein;PDBTitle: crystal structure of rbcx from thermosynechococcus elongatus
16 d1yx0a1
6.9
28
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT17 c2penE_
6.6
18
PDB header: chaperoneChain: E: PDB Molecule: orf134;PDBTitle: crystal structure of rbcx, crystal form i
18 d2peqa1
6.5
18
Fold: RbcX-likeSuperfamily: RbcX-likeFamily: RbcX-like19 c2g9mB_
6.5
18
PDB header: electron transportChain: B: PDB Molecule: phycoerythrin;PDBTitle: crystal structure of the pigment protein phycoerythrin from2 cyanobacterium at 2.6a resolution
20 c2lkyA_
5.9
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of msmeg_1053, the second duf3349 annotated protein2 in the genome of mycobacterium smegmatis, seattle structural genomics3 center for infectious disease target mysma.17112.b
21 d2py8a1
not modelled
5.9
18
Fold: RbcX-likeSuperfamily: RbcX-likeFamily: RbcX-like22 c3n4sC_
not modelled
5.7
33
PDB header: replicationChain: C: PDB Molecule: monopolin complex subunit csm1;PDBTitle: structure of csm1 c-terminal domain, p21212 form
23 c3lisB_
not modelled
5.6
11
PDB header: transcriptionChain: B: PDB Molecule: csp231i c protein;PDBTitle: crystal structure of the restriction-modification controller protein2 c.csp231i (monoclinic form)
24 c2r7hA_
not modelled
5.5
22
PDB header: transferaseChain: A: PDB Molecule: putative d-alanine n-acetyltransferase of gnat family;PDBTitle: crystal structure of a putative acetyltransferase of the gnat family2 (dde_3044) from desulfovibrio desulfuricans subsp. at 1.85 a3 resolution
25 c2py8B_
not modelled
5.5
18
PDB header: chaperoneChain: B: PDB Molecule: hypothetical protein rbcx;PDBTitle: rbcx
26 d1qaza_
not modelled
5.3
29
Fold: alpha/alpha toroidSuperfamily: Chondroitin AC/alginate lyaseFamily: Alginate lyase A1-III