| 1 |
|
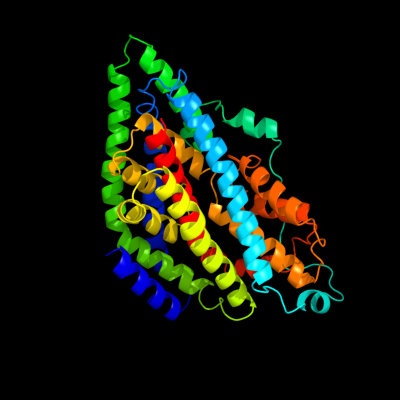
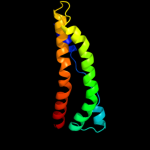
PDB 2nww chain A domain 1
Region: 12 - 402
Aligned: 382
Modelled: 391
Confidence: 100.0%
Identity: 31%
Fold: Proton glutamate symport protein
Superfamily: Proton glutamate symport protein
Family: Proton glutamate symport protein
Phyre2
| 2 |
|
PDB 1a77 chain A domain 1
Region: 324 - 362
Aligned: 38
Modelled: 39
Confidence: 42.4%
Identity: 18%
Fold: SAM domain-like
Superfamily: 5' to 3' exonuclease, C-terminal subdomain
Family: 5' to 3' exonuclease, C-terminal subdomain
Phyre2
| 3 |
|
PDB 1mc8 chain A domain 1
Region: 324 - 364
Aligned: 40
Modelled: 41
Confidence: 23.3%
Identity: 18%
Fold: SAM domain-like
Superfamily: 5' to 3' exonuclease, C-terminal subdomain
Family: 5' to 3' exonuclease, C-terminal subdomain
Phyre2
| 4 |
|
PDB 1ul1 chain X domain 1
Region: 324 - 364
Aligned: 39
Modelled: 41
Confidence: 21.2%
Identity: 15%
Fold: SAM domain-like
Superfamily: 5' to 3' exonuclease, C-terminal subdomain
Family: 5' to 3' exonuclease, C-terminal subdomain
Phyre2
| 5 |
|
PDB 1b43 chain A domain 1
Region: 324 - 362
Aligned: 38
Modelled: 39
Confidence: 18.4%
Identity: 16%
Fold: SAM domain-like
Superfamily: 5' to 3' exonuclease, C-terminal subdomain
Family: 5' to 3' exonuclease, C-terminal subdomain
Phyre2
| 6 |
|
PDB 1iwg chain A domain 8
Region: 149 - 251
Aligned: 103
Modelled: 103
Confidence: 16.5%
Identity: 13%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 7 |
|
PDB 3ah5 chain E
Region: 278 - 302
Aligned: 25
Modelled: 25
Confidence: 15.0%
Identity: 12%
PDB header:transferase
Chain: E: PDB Molecule:thymidylate synthase thyx;
PDBTitle: crystal structure of flavin dependent thymidylate synthase thyx from2 helicobacter pylori complexed with fad and dump
Phyre2
| 8 |
|
PDB 1rxw chain A domain 1
Region: 324 - 362
Aligned: 37
Modelled: 39
Confidence: 13.8%
Identity: 19%
Fold: SAM domain-like
Superfamily: 5' to 3' exonuclease, C-terminal subdomain
Family: 5' to 3' exonuclease, C-terminal subdomain
Phyre2
| 9 |
|
PDB 1xo1 chain A domain 1
Region: 325 - 369
Aligned: 43
Modelled: 45
Confidence: 11.5%
Identity: 21%
Fold: SAM domain-like
Superfamily: 5' to 3' exonuclease, C-terminal subdomain
Family: 5' to 3' exonuclease, C-terminal subdomain
Phyre2
| 10 |
|
PDB 1o26 chain A
Region: 278 - 298
Aligned: 21
Modelled: 21
Confidence: 11.3%
Identity: 19%
Fold: Thymidylate synthase-complementing protein Thy1
Superfamily: Thymidylate synthase-complementing protein Thy1
Family: Thymidylate synthase-complementing protein Thy1
Phyre2
| 11 |
|
PDB 3aqp chain B
Region: 149 - 247
Aligned: 97
Modelled: 99
Confidence: 10.0%
Identity: 11%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 12 |
|
PDB 3dl8 chain D
Region: 150 - 181
Aligned: 32
Modelled: 32
Confidence: 9.4%
Identity: 25%
PDB header:protein transport
Chain: D: PDB Molecule:sece;
PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
Phyre2
| 13 |
|
PDB 1cmw chain A domain 1
Region: 324 - 364
Aligned: 41
Modelled: 41
Confidence: 7.0%
Identity: 20%
Fold: SAM domain-like
Superfamily: 5' to 3' exonuclease, C-terminal subdomain
Family: 5' to 3' exonuclease, C-terminal subdomain
Phyre2