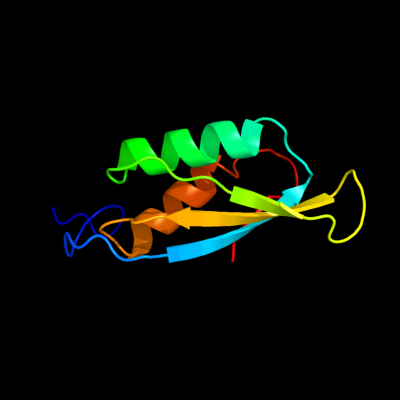
| 1 | d1rwua_
|
|
 |
100.0 |
100 |
Fold:Ferredoxin-like
Superfamily:YbeD/HP0495-like
Family:YbeD-like |
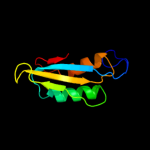
| 2 | c1rwuA_
|
|
 |
100.0 |
100 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical upf0250 protein ybed;
PDBTitle: solution structure of conserved protein ybed from e. coli
|
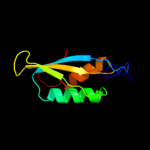
| 3 | d2joqa1
|
|
 |
99.9 |
12 |
Fold:Ferredoxin-like
Superfamily:YbeD/HP0495-like
Family:HP0495-like |
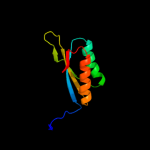
| 4 | c3ibwA_
|
|
 |
96.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:gtp pyrophosphokinase;
PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
|
| 5 | d1sc6a3
|
|
 |
82.0 |
11 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 6 | d1zpva1
|
|
 |
68.0 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:SP0238-like |
| 7 | c2jsxA_
|
|
 |
67.5 |
15 |
PDB header:chaperone
Chain: A: PDB Molecule:protein napd;
PDBTitle: solution structure of the e. coli tat proofreading2 chaperone protein napd
|
| 8 | d1u8sa1
|
|
 |
64.5 |
16 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 9 | c2cg4B_
|
|
 |
64.5 |
17 |
PDB header:transcription
Chain: B: PDB Molecule:regulatory protein asnc;
PDBTitle: structure of e.coli asnc
|
| 10 | d2fgca2
|
|
 |
57.6 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 11 | d2f1fa1
|
|
 |
56.6 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 12 | d1u69a_
|
|
 |
55.4 |
7 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:3-demethylubiquinone-9 3-methyltransferase |
| 13 | d1tsja_
|
|
 |
48.8 |
14 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:3-demethylubiquinone-9 3-methyltransferase |
| 14 | d2pc6a2
|
|
 |
46.6 |
14 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 15 | c2f1fA_
|
|
 |
43.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase isozyme iii small subunit;
PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
|
| 16 | c3n0vD_
|
|
 |
41.7 |
6 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 17 | c3k5pA_
|
|
 |
40.6 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
|
| 18 | d2qmwa2
|
|
 |
38.7 |
8 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 19 | d2cg4a2
|
|
 |
38.2 |
16 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 20 | c1u8sB_
|
|
 |
37.8 |
18 |
PDB header:transcription
Chain: B: PDB Molecule:glycine cleavage system transcriptional
PDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
|
| 21 | c3omsA_ |
|
not modelled |
29.2 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phnb protein;
PDBTitle: putative 3-demethylubiquinone-9 3-methyltransferase, phnb protein,2 from bacillus cereus.
|
| 22 | c2kxoA_ |
|
not modelled |
28.6 |
13 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division topological specificity factor;
PDBTitle: solution nmr structure of the cell division regulator mine protein2 from neisseria gonorrhoeae
|
| 23 | c2fgcA_ |
|
not modelled |
28.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase, small subunit;
PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
|
| 24 | d1u8sa2 |
|
not modelled |
26.9 |
13 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 25 | d2gz1a2 |
|
not modelled |
21.3 |
16 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 26 | c2e1cA_ |
|
not modelled |
19.6 |
18 |
PDB header:transcription/dna
Chain: A: PDB Molecule:putative hth-type transcriptional regulator ph1519;
PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
|
| 27 | c2pc6C_ |
|
not modelled |
17.0 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:probable acetolactate synthase isozyme iii (small subunit);
PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
|
| 28 | d1ygya3 |
|
not modelled |
16.6 |
3 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 29 | c2vbzA_ |
|
not modelled |
15.5 |
18 |
PDB header:dna-binding protein
Chain: A: PDB Molecule:transcriptional regulatory protein;
PDBTitle: feast or famine regulatory protein (rv3291c)from m.2 tuberculosis complexed with l-tryptophan
|
| 30 | d1u7ia_ |
|
not modelled |
14.0 |
22 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:3-demethylubiquinone-9 3-methyltransferase |
| 31 | d1phza1 |
|
not modelled |
13.1 |
7 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 32 | d1ev0a_ |
|
not modelled |
12.5 |
20 |
Fold:Cell division protein MinE topological specificity domain
Superfamily:Cell division protein MinE topological specificity domain
Family:Cell division protein MinE topological specificity domain |
| 33 | c2nyiB_ |
|
not modelled |
12.5 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:unknown protein;
PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
|
| 34 | d2aala1 |
|
not modelled |
12.2 |
14 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MSAD-like |
| 35 | d1o8ba2 |
|
not modelled |
12.2 |
16 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 36 | d1m0sa2 |
|
not modelled |
11.1 |
19 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 37 | d1uj4a2 |
|
not modelled |
10.2 |
19 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 38 | c2yx5A_ |
|
not modelled |
9.8 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0062 protein mj1593;
PDBTitle: crystal structure of methanocaldococcus jannaschii purs, one of the2 subunits of formylglycinamide ribonucleotide amidotransferase in the3 purine biosynthetic pathway
|
| 39 | c1cf2Q_ |
|
not modelled |
9.8 |
10 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:protein (glyceraldehyde-3-phosphate
PDBTitle: three-dimensional structure of d-glyceraldehyde-3-phosphate2 dehydrogenase from the hyperthermophilic archaeon3 methanothermus fervidus
|
| 40 | c3hjaB_ |
|
not modelled |
9.7 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase from borrelia burgdorferi
|
| 41 | d1lk5a2 |
|
not modelled |
9.6 |
26 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 42 | c2e1aD_ |
|
not modelled |
9.5 |
7 |
PDB header:transcription
Chain: D: PDB Molecule:75aa long hypothetical regulatory protein asnc;
PDBTitle: crystal structure of ffrp-dm1
|
| 43 | d1lp1b_ |
|
not modelled |
9.4 |
19 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 44 | d1fc2c_ |
|
not modelled |
9.4 |
19 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 45 | d2cyya2 |
|
not modelled |
9.4 |
19 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 46 | c1i1gA_ |
|
not modelled |
9.1 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator lrpa;
PDBTitle: crystal structure of the lrp-like transcriptional regulator from the2 archaeon pyrococcus furiosus
|
| 47 | d2drpa2 |
|
not modelled |
8.1 |
22 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 48 | c3mgjA_ |
|
not modelled |
7.8 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mj1480;
PDBTitle: crystal structure of the saccharop_dh_n domain of mj14802 protein from methanococcus jannaschii. northeast structural3 genomics consortium target mjr83a.
|
| 49 | c3ptyA_ |
|
not modelled |
7.7 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:arabinosyltransferase c;
PDBTitle: crystal structure of the c-terminal extracellular domain of2 mycobacterium tuberculosis embc
|
| 50 | c3i4pA_ |
|
not modelled |
7.4 |
14 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, asnc family;
PDBTitle: crystal structure of asnc family transcriptional regulator from2 agrobacterium tumefaciens
|
| 51 | c3nrbD_ |
|
not modelled |
7.3 |
6 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
| 52 | d1deeg_ |
|
not modelled |
7.1 |
13 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 53 | d2hjsa2 |
|
not modelled |
7.0 |
16 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 54 | c1vdxA_ |
|
not modelled |
6.9 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:hypothetical protein ph0099;
PDBTitle: crystal structure of a pyrococcus horikoshii protein with2 similarities to 2'5' rna-ligase
|
| 55 | c1zyiA_ |
|
not modelled |
6.9 |
5 |
PDB header:translation
Chain: A: PDB Molecule:methylosome subunit picln;
PDBTitle: solution structure of icln, a multifunctional protein2 involved in regulatory mechanisms as different as cell3 volume regulation and rna splicing
|
| 56 | c2zw2B_ |
|
not modelled |
6.5 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:putative uncharacterized protein sts178;
PDBTitle: crystal structure of formylglycinamide ribonucleotide amidotransferase2 iii from sulfolobus tokodaii (stpurs)
|
| 57 | c2cfxD_ |
|
not modelled |
6.5 |
12 |
PDB header:transcription
Chain: D: PDB Molecule:hth-type transcriptional regulator lrpc;
PDBTitle: structure of b.subtilis lrpc
|
| 58 | d2jwda1 |
|
not modelled |
6.5 |
19 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 59 | d1iuha_ |
|
not modelled |
6.3 |
16 |
Fold:LigT-like
Superfamily:LigT-like
Family:2'-5' RNA ligase LigT |
| 60 | d1udma_ |
|
not modelled |
6.1 |
15 |
Fold:Gelsolin-like
Superfamily:Actin depolymerizing proteins
Family:Cofilin-like |
| 61 | c2e7xA_ |
|
not modelled |
6.1 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:150aa long hypothetical transcriptional regulator;
PDBTitle: structure of the lrp/asnc like transcriptional regulator from2 sulfolobus tokodaii 7 complexed with its cognate ligand
|
| 62 | d2nzca1 |
|
not modelled |
6.1 |
16 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:TM1266-like |
| 63 | d1j7ma_ |
|
not modelled |
5.9 |
25 |
Fold:Kringle-like
Superfamily:Kringle-like
Family:Fibronectin type II module |
| 64 | c2k49A_ |
|
not modelled |
5.8 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0339 protein so_3888;
PDBTitle: solution nmr structure of upf0339 protein so3888 from shewanella2 oneidensis. northeast structural genomics consortium target sor190
|
| 65 | c1qo6A_ |
|
not modelled |
5.5 |
13 |
PDB header:cell adhesion protein
Chain: A: PDB Molecule:fibronectin;
PDBTitle: solution structure of a pair of modules from the2 gelatin-binding domain of fibronectin
|
| 66 | c1fllA_ |
|
not modelled |
5.5 |
22 |
PDB header:apoptosis
Chain: A: PDB Molecule:tnf receptor associated factor 3;
PDBTitle: molecular basis for cd40 signaling mediated by traf3
|
| 67 | d1mb4a2 |
|
not modelled |
5.5 |
19 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 68 | d1ks0a_ |
|
not modelled |
5.4 |
13 |
Fold:Kringle-like
Superfamily:Kringle-like
Family:Fibronectin type II module |
| 69 | c3lr4A_ |
|
not modelled |
5.4 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: periplasmic domain of the riss sensor protein from burkholderia2 pseudomallei, barium phased at low ph
|
| 70 | d1eaka5 |
|
not modelled |
5.3 |
27 |
Fold:Kringle-like
Superfamily:Kringle-like
Family:Fibronectin type II module |
| 71 | d1f08a_ |
|
not modelled |
5.3 |
15 |
Fold:Origin of replication-binding domain, RBD-like
Superfamily:Origin of replication-binding domain, RBD-like
Family:Replication initiation protein E1 |
| 72 | c3l20A_ |
|
not modelled |
5.2 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a hypothetical protein from staphylococcus aureus
|
| 73 | c2yyyB_ |
|
not modelled |
5.2 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase
|
| 74 | d1l6ja5 |
|
not modelled |
5.1 |
13 |
Fold:Kringle-like
Superfamily:Kringle-like
Family:Fibronectin type II module |