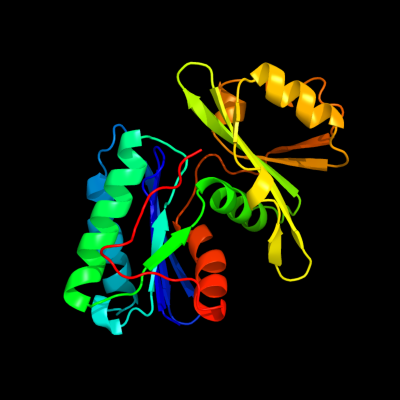
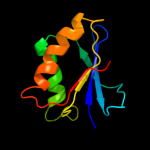
1 c1okjB_
100.0
100
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical protease yeaz;PDBTitle: crystal structure of the essential e. coli yeaz2 protein by mad method using the gadolinium complex3 "dotma"
2 c3r6mD_
100.0
55
PDB header: hydrolaseChain: D: PDB Molecule: yeaz, resuscitation promoting factor;PDBTitle: crystal structure of vibrio parahaemolyticus yeaz
3 c2a6aB_
100.0
26
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical protein tm0874;PDBTitle: crystal structure of glycoprotein endopeptidase (tm0874) from2 thermotoga maritima at 2.50 a resolution
4 d1okja1
100.0
100
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like5 d2a6aa1
100.0
21
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like6 c2ivoC_
99.9
21
PDB header: hydrolaseChain: C: PDB Molecule: up1;PDBTitle: structure of up1 protein
7 c3enoB_
99.9
22
PDB header: hydrolase/unknown functionChain: B: PDB Molecule: putative o-sialoglycoprotein endopeptidase;PDBTitle: crystal structure of pyrococcus furiosus pcc1 in complex2 with thermoplasma acidophilum kae1
8 c3en9B_
99.9
15
PDB header: hydrolaseChain: B: PDB Molecule: o-sialoglycoprotein endopeptidase/protein kinase;PDBTitle: structure of the methanococcus jannaschii kae1-bud32 fusion2 protein
9 d1okja2
99.7
100
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like10 d1huxa_
97.5
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like11 d2a6aa2
97.3
33
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like12 d1saza1
96.4
23
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Acetokinase-like13 c1sazA_
95.8
17
PDB header: transferaseChain: A: PDB Molecule: probable butyrate kinase 2;PDBTitle: membership in the askha superfamily: enzymological2 properties and crystal structure of butyrate kinase 2 from3 thermotoga maritima
14 d1hjra_
94.7
18
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: RuvC resolvase15 c2aa4B_
94.0
19
PDB header: transferaseChain: B: PDB Molecule: putative n-acetylmannosamine kinase;PDBTitle: crystal structure of escherichia coli putative n-2 acetylmannosamine kinase, new york structural genomics3 consortium
16 d1zc6a1
94.0
23
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like17 c2iikA_
93.6
11
PDB header: transferaseChain: A: PDB Molecule: 3-ketoacyl-coa thiolase, peroxisomal;PDBTitle: crystal structure of human peroxisomal acetyl-coa acyl transferase 12 (acaa1)
18 c2wuaA_
93.6
11
PDB header: transferaseChain: A: PDB Molecule: acetoacetyl coa thiolase;PDBTitle: structure of the peroxisomal 3-ketoacyl-coa thiolase from2 sunflower
19 c2qm1D_
93.5
15
PDB header: transferaseChain: D: PDB Molecule: glucokinase;PDBTitle: crystal structure of glucokinase from enterococcus faecalis
20 d1iv0a_
93.5
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX21 d1ulqa1
not modelled
93.4
8
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related22 c3goaA_
not modelled
92.9
11
PDB header: transferaseChain: A: PDB Molecule: 3-ketoacyl-coa thiolase;PDBTitle: crystal structure of the salmonella typhimurium fada 3-2 ketoacyl-coa thiolase
23 d1m3ka1
not modelled
92.8
13
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related24 d1vhxa_
not modelled
92.6
20
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX25 d1nu0a_
not modelled
92.2
13
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX26 c1ulqD_
not modelled
91.8
8
PDB header: transferaseChain: D: PDB Molecule: putative acetyl-coa acetyltransferase;PDBTitle: crystal structure of tt0182 from thermus thermophilus hb8
27 c3gg4B_
not modelled
91.7
16
PDB header: transferaseChain: B: PDB Molecule: glycerol kinase;PDBTitle: the crystal structure of glycerol kinase from yersinia2 pseudotuberculosis
28 d2a9va1
not modelled
91.7
16
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)29 c2vu2D_
not modelled
91.4
13
PDB header: transferaseChain: D: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: biosynthetic thiolase from z. ramigera. complex with s-2 pantetheine-11-pivalate.
30 c2zf5O_
not modelled
91.2
19
PDB header: transferaseChain: O: PDB Molecule: glycerol kinase;PDBTitle: crystal structure of highly thermostable glycerol kinase from a2 hyperthermophilic archaeon
31 c3hz6A_
not modelled
90.6
23
PDB header: transferaseChain: A: PDB Molecule: xylulokinase;PDBTitle: crystal structure of xylulokinase from chromobacterium violaceum
32 c3ss6B_
not modelled
90.4
10
PDB header: transferaseChain: B: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: crystal structure of the bacillus anthracis acetyl-coa2 acetyltransferase
33 d1wdkc1
not modelled
88.7
12
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related34 d1afwa1
not modelled
88.6
10
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related35 c2ibyD_
not modelled
88.4
15
PDB header: transferaseChain: D: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: crystallographic and kinetic studies of human mitochondrial2 acetoacetyl-coa thiolase (t2): the importance of potassium and3 chloride for its structure and function
36 c1z6rC_
not modelled
88.4
12
PDB header: transcriptionChain: C: PDB Molecule: mlc protein;PDBTitle: crystal structure of mlc from escherichia coli
37 c3ezwD_
not modelled
88.3
22
PDB header: transferaseChain: D: PDB Molecule: glycerol kinase;PDBTitle: crystal structure of a hyperactive escherichia coli glycerol kinase2 mutant gly230 --> asp obtained using microfluidic crystallization3 devices
38 c1zc6A_
not modelled
88.2
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: probable n-acetylglucosamine kinase;PDBTitle: crystal structure of putative n-acetylglucosamine kinase from2 chromobacterium violaceum. northeast structural genomics target3 cvr23.
39 c2dpnB_
not modelled
88.0
27
PDB header: transferaseChain: B: PDB Molecule: glycerol kinase;PDBTitle: crystal structure of the glycerol kinase from thermus2 thermophilus hb8
40 c2c7yB_
not modelled
88.0
8
PDB header: transferaseChain: B: PDB Molecule: 3-ketoacyl-coa thiolase 2;PDBTitle: plant enzyme
41 c1glbG_
not modelled
86.0
22
PDB header: phosphotransferaseChain: G: PDB Molecule: glycerol kinase;PDBTitle: structure of the regulatory complex of escherichia coli iiiglc with2 glycerol kinase
42 c2d3mA_
not modelled
85.9
12
PDB header: transferaseChain: A: PDB Molecule: pentaketide chromone synthase;PDBTitle: pentaketide chromone synthase complexed with coenzyme a
43 c1keeH_
not modelled
85.6
19
PDB header: ligaseChain: H: PDB Molecule: carbamoyl-phosphate synthetase small chain;PDBTitle: inactivation of the amidotransferase activity of carbamoyl phosphate2 synthetase by the antibiotic acivicin
44 d1ka9h_
not modelled
85.2
27
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)45 d1a9xb2
not modelled
85.2
19
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)46 c1ee0A_
not modelled
85.1
10
PDB header: transferaseChain: A: PDB Molecule: 2-pyrone synthase;PDBTitle: 2-pyrone synthase complexed with acetoacetyl-coa
47 c3eo3B_
not modelled
85.0
14
PDB header: isomerase, transferaseChain: B: PDB Molecule: bifunctional udp-n-acetylglucosamine 2-epimerase/n-PDBTitle: crystal structure of the n-acetylmannosamine kinase domain of human2 gne protein
48 c3g25B_
not modelled
84.9
23
PDB header: transferaseChain: B: PDB Molecule: glycerol kinase;PDBTitle: 1.9 angstrom crystal structure of glycerol kinase (glpk) from2 staphylococcus aureus in complex with glycerol.
49 d2p3ra1
not modelled
84.9
22
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Glycerol kinase50 c2ap1A_
not modelled
84.8
9
PDB header: transferaseChain: A: PDB Molecule: putative regulator protein;PDBTitle: crystal structure of the putative regulatory protein
51 c3flcX_
not modelled
84.5
20
PDB header: transferaseChain: X: PDB Molecule: glycerol kinase;PDBTitle: crystal structure of the his-tagged h232r mutant of glycerol kinase2 from enterococcus casseliflavus with glycerol
52 c3tsuA_
not modelled
84.2
14
PDB header: transferaseChain: A: PDB Molecule: transcriptional regulatory protein;PDBTitle: crystal structure of e. coli hypf with amp-pnp and carbamoyl phosphate
53 c2d4wA_
not modelled
84.2
25
PDB header: transferaseChain: A: PDB Molecule: glycerol kinase;PDBTitle: crystal structure of glycerol kinase from cellulomonas sp.2 nt3060
54 c2h3gX_
not modelled
83.6
15
PDB header: biosynthetic proteinChain: X: PDB Molecule: biosynthetic protein;PDBTitle: structure of the type iii pantothenate kinase (coax) from bacillus2 anthracis
55 d1g99a1
not modelled
82.6
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Acetokinase-like56 d3eeqa1
not modelled
82.4
10
Fold: CobE/GbiG C-terminal domain-likeSuperfamily: CobE/GbiG C-terminal domain-likeFamily: CobE/GbiG C-terminal domain-like57 c1afwB_
not modelled
82.3
14
PDB header: thiolaseChain: B: PDB Molecule: 3-ketoacetyl-coa thiolase;PDBTitle: the 1.8 angstrom crystal structure of the dimeric2 peroxisomal thiolase of saccharomyces cerevisiae
58 d3bzka5
not modelled
81.9
18
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like59 d1u6ea2
not modelled
81.8
9
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like60 c3ifrB_
not modelled
81.3
20
PDB header: transferaseChain: B: PDB Molecule: carbohydrate kinase, fggy;PDBTitle: the crystal structure of xylulose kinase from rhodospirillum rubrum
61 d1bi5a1
not modelled
80.8
16
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like62 c2qnxA_
not modelled
80.4
15
PDB header: transferaseChain: A: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3;PDBTitle: crystal structure of the complex between the mycobacterium beta-2 ketoacyl-acyl carrier protein synthase iii (fabh) and 11-3 [(decyloxycarbonyl)dithio]-undecanoic acid
63 c2e2pA_
not modelled
80.0
13
PDB header: transferaseChain: A: PDB Molecule: hexokinase;PDBTitle: crystal structure of sulfolobus tokodaii hexokinase in2 complex with adp
64 c3eeqB_
not modelled
79.7
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative cobalamin biosynthesis protein gPDBTitle: crystal structure of a putative cobalamin biosynthesis2 protein g homolog from sulfolobus solfataricus
65 d1ee0a2
not modelled
79.3
13
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like66 c2w40C_
not modelled
79.0
18
PDB header: transferaseChain: C: PDB Molecule: glycerol kinase, putative;PDBTitle: crystal structure of plasmodium falciparum glycerol kinase2 with bound glycerol
67 d1ub7a2
not modelled
79.0
9
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like68 d2ch5a2
not modelled
78.7
20
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like69 c2p0uB_
not modelled
78.2
15
PDB header: transferaseChain: B: PDB Molecule: stilbenecarboxylate synthase 2;PDBTitle: crystal structure of marchantia polymorpha stilbenecarboxylate2 synthase 2 (stcs2)
70 c3djcA_
not modelled
77.7
13
PDB header: transferaseChain: A: PDB Molecule: type iii pantothenate kinase;PDBTitle: crystal structure of pantothenate kinase from legionella pneumophila
71 d1mzja2
not modelled
77.5
18
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like72 c2d3tC_
not modelled
77.4
12
PDB header: lyase, oxidoreductase/transferaseChain: C: PDB Molecule: 3-ketoacyl-coa thiolase;PDBTitle: fatty acid beta-oxidation multienzyme complex from2 pseudomonas fragi, form v
73 c1u0mA_
not modelled
77.3
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative polyketide synthase;PDBTitle: crystal structure of 1,3,6,8-tetrahydroxynaphthalene synthase (thns)2 from streptomyces coelicolor a3(2): a bacterial type iii polyketide3 synthase (pks) provides insights into enzymatic control of reactive4 polyketide intermediates
74 d1u0ma2
not modelled
77.3
9
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like75 c2cgkB_
not modelled
76.9
11
PDB header: transferaseChain: B: PDB Molecule: l-rhamnulose kinase;PDBTitle: crystal structure of l-rhamnulose kinase from escherichia2 coli in an open uncomplexed conformation.
76 d1u0ua2
not modelled
76.1
10
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like77 c3i8bA_
not modelled
76.1
25
PDB header: transferaseChain: A: PDB Molecule: xylulose kinase;PDBTitle: the crystal structure of xylulose kinase from2 bifidobacterium adolescentis
78 c2ch5D_
not modelled
75.7
17
PDB header: transferaseChain: D: PDB Molecule: nagk protein;PDBTitle: crystal structure of human n-acetylglucosamine kinase in2 complex with n-acetylglucosamine
79 c3aleB_
not modelled
75.1
16
PDB header: transferaseChain: B: PDB Molecule: os07g0271500 protein;PDBTitle: a type iii polyketide synthase that produces diarylheptanoid
80 d1hnja2
not modelled
74.3
10
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like81 c1z05A_
not modelled
73.2
15
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, rok family;PDBTitle: crystal structure of the rok family transcriptional regulator, homolog2 of e.coli mlc protein.
82 d1k9vf_
not modelled
72.9
23
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)83 c3euoB_
not modelled
71.2
10
PDB header: transferaseChain: B: PDB Molecule: type iii pentaketide synthase;PDBTitle: crystal structure of a fungal type iii polyketide synthase,2 oras
84 d1mzja1
not modelled
70.9
12
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like85 d1bi5a2
not modelled
70.8
21
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like86 c3oitB_
not modelled
70.6
13
PDB header: transferaseChain: B: PDB Molecule: os07g0271500 protein;PDBTitle: crystal structure of curcuminoid synthase cus from oryza sativa
87 c3fk5A_
not modelled
70.0
13
PDB header: transferaseChain: A: PDB Molecule: 3-oxoacyl-synthase iii;PDBTitle: crystal structure of 3-oxoacyl-(acyl carrier protein)2 synthase iii, fabh (xoo4209) from xanthomonas oryzae pv.3 oryzae kacc10331
88 c3il3A_
not modelled
69.1
6
PDB header: transferaseChain: A: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3;PDBTitle: structure of haemophilus influenzae fabh
89 d1z6ra2
not modelled
69.1
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK90 c2ad5B_
not modelled
69.1
28
PDB header: ligaseChain: B: PDB Molecule: ctp synthase;PDBTitle: mechanisms of feedback regulation and drug resistance of ctp2 synthetases: structure of the e. coli ctps/ctp complex at 2.8-3 angstrom resolution.
91 c3il5D_
not modelled
69.0
11
PDB header: transferaseChain: D: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3;PDBTitle: structure of e. faecalis fabh in complex with 2-({4-bromo-3-2 [(diethylamino)sulfonyl]benzoyl}amino)benzoic acid
92 c1xetD_
not modelled
68.6
15
PDB header: transferaseChain: D: PDB Molecule: dihydropinosylvin synthase;PDBTitle: crystal structure of stilbene synthase from pinus2 sylvestris, complexed with methylmalonyl coa
93 c3nvaB_
not modelled
68.5
28
PDB header: ligaseChain: B: PDB Molecule: ctp synthase;PDBTitle: dimeric form of ctp synthase from sulfolobus solfataricus
94 c1vcnA_
not modelled
67.9
24
PDB header: ligaseChain: A: PDB Molecule: ctp synthetase;PDBTitle: crystal structure of t.th. hb8 ctp synthetase complex with sulfate2 anion
95 c3ov3A_
not modelled
66.6
19
PDB header: transferaseChain: A: PDB Molecule: curcumin synthase;PDBTitle: g211f mutant of curcumin synthase 1 from curcuma longa
96 d1z05a3
not modelled
66.3
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK97 c3e1hA_
not modelled
65.6
12
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a type iii polyketide synthase2 pksiiinc from neurospora crassa
98 c1tqyD_
not modelled
65.1
14
PDB header: transferaseChain: D: PDB Molecule: actinorhodin polyketide putative beta-ketoacyl synthase 2;PDBTitle: the actinorhodin ketosynthase/chain length factor
99 c1cmlA_
not modelled
64.9
12
PDB header: transferaseChain: A: PDB Molecule: protein (chalcone synthase);PDBTitle: chalcone synthase from alfalfa complexed with malonyl-coa
100 c2gyoB_
not modelled
64.5
6
PDB header: transferaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3;PDBTitle: methanethiol-cys 112 inhibition complex of e. coli ketoacyl synthase2 iii (fabh) and coenzyme a
101 c1e5mA_
not modelled
64.3
18
PDB header: condensing enzymeChain: A: PDB Molecule: beta ketoacyl acyl carrier protein synthase ii;PDBTitle: beta ketoacyl acyl carrier protein synthase ii (kasii) from2 synechocystis sp.
102 d3bexa1
not modelled
63.7
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: CoaX-like103 d1ub7a1
not modelled
62.0
16
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like104 c2h84A_
not modelled
61.9
15
PDB header: biosynthetic protein, transferaseChain: A: PDB Molecule: steely1;PDBTitle: crystal structure of the c-terminal type iii polyketide synthase (pks2 iii) domain of 'steely1' (a type i/iii pks hybrid from dictyostelium)
105 d2ewsa1
not modelled
61.4
16
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Fumble-like106 d1u0ua1
not modelled
61.3
14
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like107 d1afwa2
not modelled
61.3
14
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related108 c3gbtA_
not modelled
61.0
20
PDB header: transferaseChain: A: PDB Molecule: gluconate kinase;PDBTitle: crystal structure of gluconate kinase from lactobacillus acidophilus
109 d1wdkc2
not modelled
60.9
22
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Thiolase-related110 c2ebdB_
not modelled
60.5
9
PDB header: transferaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3;PDBTitle: crystal structure of 3-oxoacyl-[acyl-carrier-protein] synthase iii2 from aquifex aeolicus vf5
111 c3uowB_
not modelled
60.4
19
PDB header: ligaseChain: B: PDB Molecule: gmp synthetase;PDBTitle: crystal structure of pf10_0123, a gmp synthetase from plasmodium2 falciparum
112 c1mzjB_
not modelled
60.1
15
PDB header: transferaseChain: B: PDB Molecule: beta-ketoacylsynthase iii;PDBTitle: crystal structure of the priming beta-ketosynthase from the2 r1128 polyketide biosynthetic pathway
113 c1jvnB_
not modelled
59.5
17
PDB header: transferaseChain: B: PDB Molecule: bifunctional histidine biosynthesis protein hishf;PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
114 d1qdlb_
not modelled
59.0
23
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)115 c3s3lB_
not modelled
58.6
9
PDB header: transferaseChain: B: PDB Molecule: cerj;PDBTitle: crystal structure of cerj from streptomyces tendae
116 c1zowB_
not modelled
58.5
10
PDB header: transferaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase iii;PDBTitle: crystal structure of s. aureus fabh, beta-ketoacyl carrier protein2 synthase iii
117 d1i1qb_
not modelled
57.8
23
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)118 c2w7tA_
not modelled
56.7
24
PDB header: ligaseChain: A: PDB Molecule: putative cytidine triphosphate synthase;PDBTitle: trypanosoma brucei ctps - glutaminase domain with bound2 acivicin
119 d2aa4a1
not modelled
56.7
16
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK120 d2ap1a2
not modelled
56.7
11
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK