1 c3on0D_
100.0
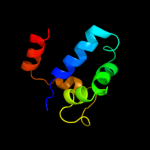
39
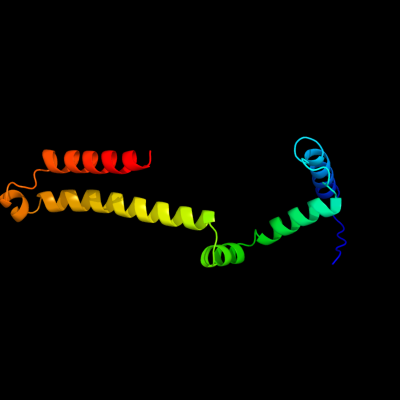
PDB header: dna binding protein/dnaChain: D: PDB Molecule: protein tram;PDBTitle: crystal structure of the ped208 tram-sbma complex
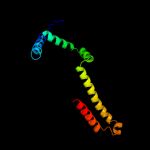
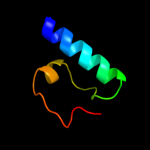
2 d2g7oa1
99.9
100
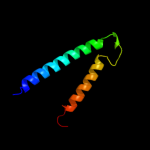
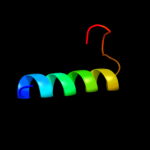
Fold: TraM-likeSuperfamily: TraM-likeFamily: TraM-like3 d1dp3a_
99.9
64
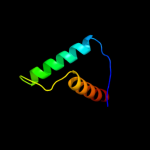
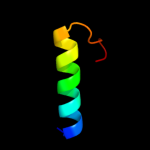
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: DNA-binding domain (fragment?) of the TraM protein4 c3omyB_
99.9
48
PDB header: dna binding proteinChain: B: PDB Molecule: protein tram;PDBTitle: crystal structure of the ped208 tram n-terminal domain
5 d1tlha_
42.5
15
Fold: Anti-sigma factor AsiASuperfamily: Anti-sigma factor AsiAFamily: Anti-sigma factor AsiA6 c2qwtA_
23.8
20
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: crystal structure of the tetr transcription regulatory2 protein from mycobacterium vanbaalenii
7 c2jvdA_
15.1
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0291 protein ynzc;PDBTitle: solution nmr structure of the folded n-terminal fragment of2 upf0291 protein ynzc from bacillus subtilis. northeast3 structural genomics target sr384-1-46
8 c3bhpA_
14.7
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0291 protein ynzc;PDBTitle: crystal structure of upf0291 protein ynzc from bacillus2 subtilis at resolution 2.0 a. northeast structural3 genomics consortium target sr384
9 c2l53B_
14.4
36
PDB header: ca-binding protein/proton transportChain: B: PDB Molecule: voltage-gated sodium channel type v alpha isoform bPDBTitle: solution nmr structure of apo-calmodulin in complex with the iq motif2 of human cardiac sodium channel nav1.5
10 d1oysa2
13.2
25
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like11 d1x2la1
11.5
18
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain12 c3bpqC_
11.1
32
PDB header: toxinChain: C: PDB Molecule: antitoxin relb3;PDBTitle: crystal structure of relb-rele antitoxin-toxin complex from2 methanococcus jannaschii
13 c2r17C_
10.8
14
PDB header: protein transportChain: C: PDB Molecule: vacuolar protein sorting-associated protein 35;PDBTitle: functional architecture of the retromer cargo-recognition complex
14 c3r9lA_
10.4
13
PDB header: transferaseChain: A: PDB Molecule: nucleoside diphosphate kinase;PDBTitle: crystal structure of nucleoside diphosphate kinase from giardia2 lamblia featuring a disordered dinucleotide binding site
15 d2v7qi1
10.2
23
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Epsilon subunit of mitochondrial F1F0-ATP synthaseFamily: Epsilon subunit of mitochondrial F1F0-ATP synthase16 c2jdqB_
9.9
25
PDB header: protein transportChain: B: PDB Molecule: importin alpha-1 subunit;PDBTitle: c-terminal domain of influenza a virus polymerase pb22 subunit in complex with human importin alpha5
17 c2cp8A_
8.4
29
PDB header: protein bindingChain: A: PDB Molecule: next to brca1 gene 1 protein;PDBTitle: solution structure of the rsgi ruh-046, a uba domain from2 human next to brca1 gene 1 protein (kiaa0049 protein)3 r923h variant
18 d1zkda1
8.4
28
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: RPA4359-like19 d1l7da2
7.3
16
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: L-alanine dehydrogenase-like20 c2kxwB_
7.2
36
PDB header: calcium-binding protein/metal transportChain: B: PDB Molecule: sodium channel protein type 2 subunit alpha;PDBTitle: structure of the c-domain fragment of apo calmodulin bound to the iq2 motif of nav1.2
21 c3mtuE_
not modelled
7.1
28
PDB header: contractile proteinChain: E: PDB Molecule: head morphogenesis protein, tropomyosin alpha-1 chain;PDBTitle: structure of the tropomyosin overlap complex from chicken smooth2 muscle
22 d1f7ca_
not modelled
6.9
14
Fold: GTPase activation domain, GAPSuperfamily: GTPase activation domain, GAPFamily: BCR-homology GTPase activation domain (BH-domain)23 c1f7cA_
not modelled
6.9
14
PDB header: signaling proteinChain: A: PDB Molecule: rhogap protein;PDBTitle: crystal structure of the bh domain from graf, the gtpase2 regulator associated with focal adhesion kinase
24 c3izcR_
not modelled
6.4
24
PDB header: ribosomeChain: R: PDB Molecule: 60s ribosomal protein rpl18 (l18e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
25 d1nhkl_
not modelled
6.2
25
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK26 c3pk1A_
not modelled
6.2
20
PDB header: apoptosis/apoptosis regulatorChain: A: PDB Molecule: induced myeloid leukemia cell differentiation protein mcl-PDBTitle: crystal structure of mcl-1 in complex with the baxbh3 domain
27 d1kx5b_
not modelled
6.0
21
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones28 d2o4aa1
not modelled
6.0
23
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain29 d1xiqa_
not modelled
6.0
13
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK30 c2hurF_
not modelled
5.9
27
PDB header: signaling protein,transferaseChain: F: PDB Molecule: nucleoside diphosphate kinase;PDBTitle: escherichia coli nucleoside diphosphate kinase
31 c2imuA_
not modelled
5.8
22
PDB header: viral proteinChain: A: PDB Molecule: structural polyprotein (pp) p1;PDBTitle: nmr structure of pep46 from the infectious bursal disease2 virus (ibdv) in dodecylphosphocholin (dpc).
32 d1ehwa_
not modelled
5.7
20
Fold: Ferredoxin-likeSuperfamily: Nucleoside diphosphate kinase, NDKFamily: Nucleoside diphosphate kinase, NDK33 c3knyA_
not modelled
5.5
39
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein bt_3535;PDBTitle: crystal structure of a two domain protein with unknown function2 (bt_3535) from bacteroides thetaiotaomicron vpi-5482 at 2.60 a3 resolution
34 c2o01J_
not modelled
5.4
63
PDB header: photosynthesisChain: J: PDB Molecule: photosystem i reaction center subunit ix;PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution