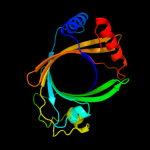
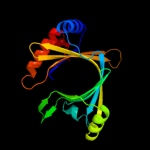
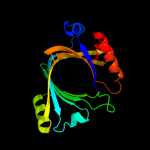
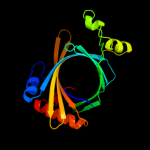
1 c3e0sA_
100.0
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from2 chlorobium tepidum
2 c2gfgB_
100.0
24
PDB header: unknown functionChain: B: PDB Molecule: bh2851;PDBTitle: crystal structure of a putative adenylate cyclase (bh2851) from2 bacillus halodurans at 2.12 a resolution
3 d2jmua1
100.0
19
Fold: CYTH-like phosphatasesSuperfamily: CYTH-like phosphatasesFamily: CYTH domain4 c3sy3D_
100.0
23
PDB header: lyaseChain: D: PDB Molecule: gbaa_1210 protein;PDBTitle: gbaa_1210 protein, a putative adenylate cyclase, from bacillus2 anthracis
5 c2fjtA_
99.8
18
PDB header: lyaseChain: A: PDB Molecule: adenylyl cyclase class iv;PDBTitle: adenylyl cyclase class iv from yersinia pestis
6 d2acaa1
99.8
16
Fold: CYTH-like phosphatasesSuperfamily: CYTH-like phosphatasesFamily: CYTH domain7 c2dc4A_
99.8
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 165aa long hypothetical protein;PDBTitle: structure of ph1012 protein from pyrococcus horikoshii ot3
8 d1yema_
99.7
17
Fold: CYTH-like phosphatasesSuperfamily: CYTH-like phosphatasesFamily: CYTH domain9 c2eenA_
99.7
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ph1819;PDBTitle: structure of ph1819 protein from pyrococcus horikoshii ot3
10 c3g3rA_
98.5
16
PDB header: biosynthetic proteinChain: A: PDB Molecule: vacuolar transporter chaperone 4;PDBTitle: crystal structure of a eukaryotic polyphosphate polymerase2 in complex with appnhp-mn2+
11 c3g3oA_
98.3
17
PDB header: biosynthetic proteinChain: A: PDB Molecule: vacuolar transporter chaperone 2;PDBTitle: crystal structure of the cytoplasmic tunnel domain in yeast2 vtc2p
12 d2fbla1
97.2
28
Fold: CYTH-like phosphatasesSuperfamily: CYTH-like phosphatasesFamily: CYTH domain13 c3oq0D_
21.0
43
PDB header: cell cycleChain: D: PDB Molecule: dbf4;PDBTitle: crystal structure of motif n of saccharomyces cerevisiae dbf4
14 d1b77a2
9.5
20
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor15 c2y3yC_
9.4
26
PDB header: transcriptionChain: C: PDB Molecule: putative nickel-responsive regulator;PDBTitle: holo-ni(ii) hpnikr is a symmetric tetramer containing four2 canonic square-planar ni(ii) ions at physiological ph
16 c2aghA_
8.9
63
PDB header: transcriptionChain: A: PDB Molecule: myb proto-oncogene protein;PDBTitle: structural basis for cooperative transcription factor2 binding to the cbp coactivator
17 d1czda2
8.8
20
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor18 d1nh1a_
8.4
15
Fold: Antivirulence factorSuperfamily: Antivirulence factorFamily: Antivirulence factor19 c1nh1A_
8.4
15
PDB header: avirulence proteinChain: A: PDB Molecule: avirulence b protein;PDBTitle: crystal structure of the type iii effector avrb from2 pseudomonas syringae.
20 c2y9xG_
7.8
28
PDB header: oxidoreductaseChain: G: PDB Molecule: lectin-like fold protein;PDBTitle: crystal structure of ppo3, a tyrosinase from agaricus bisporus, in2 deoxy-form that contains additional unknown lectin-like subunit,3 with inhibitor tropolone
21 c1sb0B_
not modelled
7.6
63
PDB header: transcriptionChain: B: PDB Molecule: protein c-myb;PDBTitle: solution structure of the kix domain of cbp bound to the2 transactivation domain of c-myb
22 d1n8ia_
not modelled
7.4
22
Fold: TIM beta/alpha-barrelSuperfamily: Malate synthase GFamily: Malate synthase G23 c3ectA_
not modelled
7.4
12
PDB header: transferaseChain: A: PDB Molecule: hexapeptide-repeat containing-acetyltransferase;PDBTitle: crystal structure of the hexapeptide-repeat containing-2 acetyltransferase vca0836 from vibrio cholerae
24 c2imuA_
not modelled
7.2
28
PDB header: viral proteinChain: A: PDB Molecule: structural polyprotein (pp) p1;PDBTitle: nmr structure of pep46 from the infectious bursal disease2 virus (ibdv) in dodecylphosphocholin (dpc).
25 d2oyza1
not modelled
7.1
33
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: VPA0057-like26 d1qr5a_
not modelled
6.8
31
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like27 d1ka5a_
not modelled
6.8
25
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like28 c2eqeA_
not modelled
6.6
43
PDB header: hydrolaseChain: A: PDB Molecule: tumor necrosis factor, alpha-induced protein 3;PDBTitle: solution structure of the fourth a20-type zinc finger2 domain from human tumor necrosis factor, alpha-induced3 protein3
29 d2ae8a2
not modelled
6.6
16
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase30 c2bnoA_
not modelled
6.5
20
PDB header: oxidoreductaseChain: A: PDB Molecule: epoxidase;PDBTitle: the structure of hydroxypropylphosphonic acid epoxidase2 from s. wedmorenis.
31 d2ofya1
not modelled
6.3
20
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like32 d1ok7a2
not modelled
6.2
15
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase III, beta subunit33 d2nzul1
not modelled
6.1
31
Fold: HPr-likeSuperfamily: HPr-likeFamily: HPr-like34 c3eo6B_
not modelled
6.1
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein of unknown function (duf1255);PDBTitle: crystal structure of protein of unknown function (duf1255)2 (afe_2634) from acidithiobacillus ferrooxidans ncib8455 at3 0.97 a resolution
35 c3pxpA_
not modelled
6.0
20
PDB header: transcription regulatorChain: A: PDB Molecule: helix-turn-helix domain protein;PDBTitle: crystal structure of a pas and dna binding domain containing protein2 (caur_2278) from chloroflexus aurantiacus j-10-fl at 2.30 a3 resolution
36 c3hqxA_
not modelled
5.9
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0345 protein aciad0356;PDBTitle: crystal structure of protein of unknown function (duf1255,pf06865)2 from acinetobacter sp. adp1
37 c3fbnC_
not modelled
5.8
17
PDB header: transcriptionChain: C: PDB Molecule: mediator of rna polymerase ii transcription subunit 7;PDBTitle: structure of the mediator submodule med7n/31
38 c1y9qA_
not modelled
5.8
25
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, hth_3 family;PDBTitle: crystal structure of hth_3 family transcriptional regulator2 from vibrio cholerae
39 c3eusB_
not modelled
5.8
15
PDB header: dna binding proteinChain: B: PDB Molecule: dna-binding protein;PDBTitle: the crystal structure of the dna binding protein from silicibacter2 pomeroyi
40 d1w0ba_
not modelled
5.8
20
Fold: Spectrin repeat-likeSuperfamily: Alpha-hemoglobin stabilizing protein AHSPFamily: Alpha-hemoglobin stabilizing protein AHSP41 c3f3bA_
not modelled
5.7
67
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phage-like element pbsx protein xkdh;PDBTitle: structure of the phage-like element pbsx protein xkdh from2 bacillus subtilus. northeast structural genomics3 consortium target sr352.
42 d2j0oa1
not modelled
5.6
9
Fold: IpaD-likeSuperfamily: IpaD-likeFamily: IpaD-like43 c2j0oA_
not modelled
5.6
9
PDB header: cell invasionChain: A: PDB Molecule: invasin ipad;PDBTitle: shigella flexneri ipad
44 d1d8ca_
not modelled
5.6
19
Fold: TIM beta/alpha-barrelSuperfamily: Malate synthase GFamily: Malate synthase G45 c2hw2A_
not modelled
5.5
14
PDB header: transferaseChain: A: PDB Molecule: rifampin adp-ribosyl transferase;PDBTitle: crystal structure of rifampin adp-ribosyl transferase in2 complex with rifampin
46 d1xu2r_
not modelled
5.3
25
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: BAFF receptor-like47 c1xu2R_
not modelled
5.3
25
PDB header: cytokine, hormone/growth factor receptorChain: R: PDB Molecule: tumor necrosis factor receptor superfamily member 17;PDBTitle: the crystal structure of april bound to bcma
48 d1d8ia_
not modelled
5.3
23
Fold: CYTH-like phosphatasesSuperfamily: CYTH-like phosphatasesFamily: mRNA triphosphatase CET149 c1b8hA_
not modelled
5.1
21
PDB header: transferaseChain: A: PDB Molecule: dna polymerase processivity component;PDBTitle: sliding clamp, dna polymerase
50 d1z8ua1
not modelled
5.1
20
Fold: Spectrin repeat-likeSuperfamily: Alpha-hemoglobin stabilizing protein AHSPFamily: Alpha-hemoglobin stabilizing protein AHSP51 c3sr2A_
not modelled
5.1
3
PDB header: dna binding protein/protein bindingChain: A: PDB Molecule: dna repair protein xrcc4;PDBTitle: crystal structure of human xlf-xrcc4 complex
52 d1v7wa2
not modelled
5.1
5
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Glycosyltransferase family 36 N-terminal domain