| 1 |
|
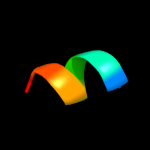
PDB 1dxz chain A
Region: 20 - 44
Aligned: 25
Modelled: 25
Confidence: 14.0%
Identity: 16%
PDB header:transmembrane protein
Chain: A: PDB Molecule:acetylcholine receptor protein, alpha chain;
PDBTitle: m2 transmembrane segment of alpha-subunit of nicotinic2 acetylcholine receptor from torpedo californica, nmr, 203 structures
Phyre2
| 2 |
|
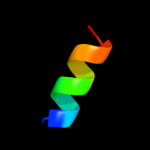
PDB 3din chain F
Region: 51 - 100
Aligned: 49
Modelled: 50
Confidence: 13.8%
Identity: 18%
PDB header:membrane protein, protein transport
Chain: F: PDB Molecule:preprotein translocase subunit secy;
PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
Phyre2
| 3 |
|
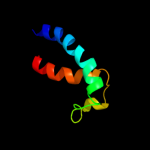
PDB 2r6g chain F domain 1
Region: 16 - 63
Aligned: 48
Modelled: 48
Confidence: 13.6%
Identity: 8%
Fold: MalF N-terminal region-like
Superfamily: MalF N-terminal region-like
Family: MalF N-terminal region-like
Phyre2
| 4 |
|
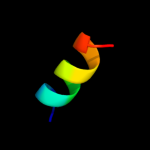
PDB 1xl7 chain A domain 2
Region: 54 - 65
Aligned: 12
Modelled: 12
Confidence: 9.8%
Identity: 17%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 5 |
|
PDB 2wwa chain C
Region: 20 - 27
Aligned: 8
Modelled: 8
Confidence: 8.8%
Identity: 63%
PDB header:ribosome
Chain: C: PDB Molecule:protein transport protein seb2;
PDBTitle: cryo-em structure of idle yeast ssh1 complex bound to the2 yeast 80s ribosome
Phyre2
| 6 |
|
PDB 1t1u chain A domain 2
Region: 54 - 65
Aligned: 12
Modelled: 12
Confidence: 8.7%
Identity: 25%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 7 |
|
PDB 3j01 chain A
Region: 43 - 100
Aligned: 57
Modelled: 58
Confidence: 8.4%
Identity: 21%
PDB header:ribosome/ribosomal protein
Chain: A: PDB Molecule:preprotein translocase secy subunit;
PDBTitle: structure of the ribosome-secye complex in the membrane environment
Phyre2
| 8 |
|
PDB 1ndb chain A domain 2
Region: 54 - 65
Aligned: 12
Modelled: 12
Confidence: 8.0%
Identity: 33%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 9 |
|
PDB 1nm8 chain A domain 2
Region: 54 - 65
Aligned: 12
Modelled: 12
Confidence: 7.0%
Identity: 25%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 10 |
|
PDB 2y69 chain W
Region: 72 - 109
Aligned: 35
Modelled: 38
Confidence: 6.9%
Identity: 11%
PDB header:electron transport
Chain: W: PDB Molecule:cytochrome c oxidase polypeptide 7a1;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 11 |
|
PDB 3dl8 chain H
Region: 43 - 100
Aligned: 58
Modelled: 58
Confidence: 6.3%
Identity: 17%
PDB header:protein transport
Chain: H: PDB Molecule:preprotein translocase subunit secy;
PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
Phyre2
| 12 |
|
PDB 1oed chain C
Region: 20 - 104
Aligned: 80
Modelled: 85
Confidence: 5.9%
Identity: 21%
Fold: Neurotransmitter-gated ion-channel transmembrane pore
Superfamily: Neurotransmitter-gated ion-channel transmembrane pore
Family: Neurotransmitter-gated ion-channel transmembrane pore
Phyre2
| 13 |
|
PDB 2ksr chain A
Region: 20 - 60
Aligned: 41
Modelled: 41
Confidence: 5.7%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:neuronal acetylcholine receptor subunit beta-2;
PDBTitle: nmr structures of tm domain of the n-acetylcholine receptor b2 subunit
Phyre2
| 14 |
|
PDB 3eam chain B
Region: 20 - 100
Aligned: 81
Modelled: 81
Confidence: 5.4%
Identity: 15%
PDB header:membrane protein, transport protein
Chain: B: PDB Molecule:glr4197 protein;
PDBTitle: an open-pore structure of a bacterial pentameric ligand-2 gated ion channel
Phyre2
| 15 |
|
PDB 1rkl chain A
Region: 72 - 93
Aligned: 22
Modelled: 22
Confidence: 5.4%
Identity: 32%
Fold: Single transmembrane helix
Superfamily: Oligosaccharyltransferase subunit ost4p
Family: Oligosaccharyltransferase subunit ost4p
Phyre2