| 1 |
|
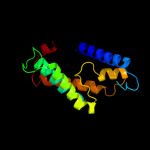
PDB 1s7b chain A
Region: 81 - 150
Aligned: 63
Modelled: 70
Confidence: 94.4%
Identity: 16%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
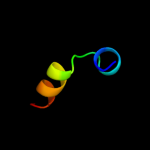
| 2 |
|
PDB 2w8a chain C
Region: 88 - 337
Aligned: 234
Modelled: 250
Confidence: 34.2%
Identity: 11%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 3 |
|
PDB 2ht2 chain B
Region: 157 - 330
Aligned: 162
Modelled: 170
Confidence: 26.0%
Identity: 11%
PDB header:membrane protein
Chain: B: PDB Molecule:h(+)/cl(-) exchange transporter clca;
PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
Phyre2
| 4 |
|
PDB 3gia chain A
Region: 4 - 338
Aligned: 324
Modelled: 335
Confidence: 21.4%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 5 |
|
PDB 2i68 chain B
Region: 99 - 150
Aligned: 39
Modelled: 52
Confidence: 18.8%
Identity: 26%
PDB header:transport protein
Chain: B: PDB Molecule:protein emre;
PDBTitle: cryo-em based theoretical model structure of transmembrane2 domain of the multidrug-resistance antiporter from e. coli3 emre
Phyre2
| 6 |
|
PDB 1ots chain A
Region: 153 - 335
Aligned: 172
Modelled: 179
Confidence: 16.3%
Identity: 10%
Fold: Clc chloride channel
Superfamily: Clc chloride channel
Family: Clc chloride channel
Phyre2
| 7 |
|
PDB 1jb0 chain L
Region: 115 - 174
Aligned: 60
Modelled: 60
Confidence: 11.6%
Identity: 13%
Fold: Photosystem I reaction center subunit XI, PsaL
Superfamily: Photosystem I reaction center subunit XI, PsaL
Family: Photosystem I reaction center subunit XI, PsaL
Phyre2
| 8 |
|
PDB 1kf6 chain D
Region: 26 - 58
Aligned: 33
Modelled: 33
Confidence: 7.7%
Identity: 30%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 9 |
|
PDB 1q90 chain B
Region: 307 - 340
Aligned: 33
Modelled: 34
Confidence: 6.8%
Identity: 15%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 10 |
|
PDB 1ymg chain A domain 1
Region: 103 - 247
Aligned: 136
Modelled: 145
Confidence: 5.7%
Identity: 13%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 11 |
|
PDB 1ymg chain A
Region: 103 - 247
Aligned: 136
Modelled: 145
Confidence: 5.7%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
Phyre2
| 12 |
|
PDB 1ycy chain A domain 1
Region: 310 - 319
Aligned: 8
Modelled: 10
Confidence: 5.4%
Identity: 63%
Fold: Sm-like fold
Superfamily: Sm-like ribonucleoproteins
Family: PF1955-like
Phyre2
| 13 |
|
PDB 2e74 chain A domain 1
Region: 307 - 340
Aligned: 33
Modelled: 34
Confidence: 5.3%
Identity: 15%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 14 |
|
PDB 3gbg chain A
Region: 309 - 327
Aligned: 19
Modelled: 19
Confidence: 5.1%
Identity: 21%
PDB header:transcription regulator
Chain: A: PDB Molecule:tcp pilus virulence regulatory protein;
PDBTitle: crystal structure of toxt from vibrio cholerae o395
Phyre2