| 1 |
|
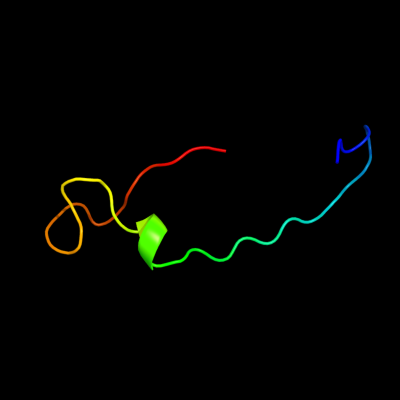
PDB 1sdd chain B domain 1
Region: 50 - 86
Aligned: 33
Modelled: 33
Confidence: 28.4%
Identity: 21%
Fold: Cupredoxin-like
Superfamily: Cupredoxins
Family: Multidomain cupredoxins
Phyre2
| 2 |
|
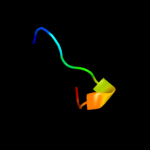
PDB 1tvt chain A
Region: 27 - 36
Aligned: 10
Modelled: 10
Confidence: 17.7%
Identity: 80%
PDB header:transcription regulation
Chain: A: PDB Molecule:transactivator protein;
PDBTitle: structure of the equine infectious anemia virus tat protein
Phyre2
| 3 |
|
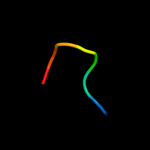
PDB 1mkf chain A
Region: 46 - 95
Aligned: 50
Modelled: 50
Confidence: 17.1%
Identity: 22%
Fold: Viral chemokine binding protein m3
Superfamily: Viral chemokine binding protein m3
Family: Viral chemokine binding protein m3
Phyre2
| 4 |
|
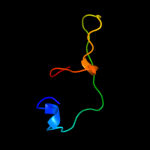
PDB 1kqh chain A
Region: 78 - 85
Aligned: 8
Modelled: 8
Confidence: 16.5%
Identity: 75%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: omega toxin-like
Family: Spider toxins
Phyre2
| 5 |
|
PDB 3nsw chain A
Region: 60 - 94
Aligned: 33
Modelled: 35
Confidence: 9.0%
Identity: 45%
PDB header:immune system
Chain: A: PDB Molecule:excretory-secretory protein 2;
PDBTitle: crystal structure of ancylostoma ceylanicum excretory-secretory2 protein 2
Phyre2
| 6 |
|
PDB 2r7t chain A
Region: 25 - 48
Aligned: 21
Modelled: 24
Confidence: 8.6%
Identity: 48%
PDB header:transferase/rna
Chain: A: PDB Molecule:rna-dependent rna polymerase;
PDBTitle: crystal structure of rotavirus sa11 vp1/rna (ugugaacc)2 complex
Phyre2
| 7 |
|
PDB 1jmu chain E
Region: 22 - 29
Aligned: 8
Modelled: 8
Confidence: 5.6%
Identity: 63%
PDB header:viral protein
Chain: E: PDB Molecule:protein mu-1;
PDBTitle: crystal structure of the reovirus mu1/sigma3 complex
Phyre2
| 8 |
|
PDB 2i1t chain A
Region: 74 - 84
Aligned: 11
Modelled: 11
Confidence: 5.5%
Identity: 55%
PDB header:toxin
Chain: A: PDB Molecule:jingzhaotoxin-3;
PDBTitle: solution structure of jingzhaotoxin-iii, a novel toxin2 inhibiting both nav and kv channels
Phyre2
| 9 |
|
PDB 1yew chain I
Region: 26 - 66
Aligned: 40
Modelled: 41
Confidence: 5.4%
Identity: 33%
PDB header:oxidoreductase, membrane protein
Chain: I: PDB Molecule:particulate methane monooxygenase, b subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
Phyre2
| 10 |
|
PDB 3rgb chain A
Region: 26 - 66
Aligned: 40
Modelled: 41
Confidence: 5.4%
Identity: 33%
PDB header:oxidoreductase
Chain: A: PDB Molecule:methane monooxygenase subunit b2;
PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
Phyre2
| 11 |
|
PDB 1kcw chain A
Region: 50 - 86
Aligned: 33
Modelled: 37
Confidence: 5.3%
Identity: 27%
PDB header:oxidoreductase
Chain: A: PDB Molecule:ceruloplasmin;
PDBTitle: x-ray crystal structure of human ceruloplasmin at 3.0 angstroms
Phyre2