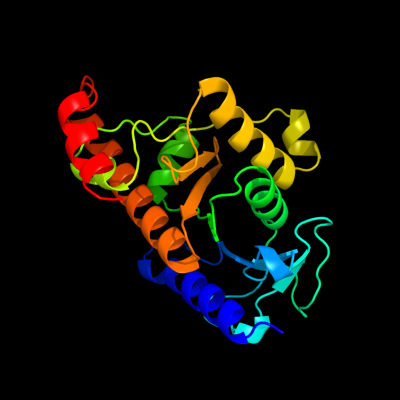
1 d1k0wa_
100.0
74
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: AraD-like aldolase/epimerase2 c3ocrA_
100.0
17
PDB header: lyaseChain: A: PDB Molecule: class ii aldolase/adducin domain protein;PDBTitle: crystal structure of aldolase ii superfamily protein from pseudomonas2 syringae
3 d1e4cp_
100.0
27
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: AraD-like aldolase/epimerase4 d1ojra_
100.0
15
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: AraD-like aldolase/epimerase5 c2z7bA_
100.0
15
PDB header: lyaseChain: A: PDB Molecule: mlr6791 protein;PDBTitle: crystal structure of mesorhizobium loti 3-hydroxy-2-methylpyridine-4,2 5-dicarboxylate decarboxylase
6 d1pvta_
100.0
22
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: AraD-like aldolase/epimerase7 c2opiB_
100.0
17
PDB header: lyaseChain: B: PDB Molecule: l-fuculose-1-phosphate aldolase;PDBTitle: crystal structure of l-fuculose-1-phosphate aldolase from bacteroides2 thetaiotaomicron
8 c2irpA_
100.0
22
PDB header: lyaseChain: A: PDB Molecule: putative aldolase class 2 protein aq_1979;PDBTitle: crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979)2 from aquifex aeolicus vf5
9 c2fk5B_
100.0
27
PDB header: lyaseChain: B: PDB Molecule: fuculose-1-phosphate aldolase;PDBTitle: crystal structure of l-fuculose-1-phosphate aldolase from thermus2 thermophilus hb8
10 c3m4rA_
100.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: structure of the n-terminal class ii aldolase domain of a conserved2 protein from thermoplasma acidophilum
11 c2cfuA_
34.7
11
PDB header: hydrolaseChain: A: PDB Molecule: sdsa1;PDBTitle: crystal structure of sdsa1, an alkylsulfatase from2 pseudomonas aeruginosa, in complex with 1-decane-sulfonic-3 acid.
12 c2yztA_
34.1
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha1756;PDBTitle: crystal structure of uncharacterized conserved protein from thermus2 thermophilus hb8
13 d2cfua2
27.0
11
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Alkylsulfatase-like14 c3kvaA_
19.2
20
PDB header: h3k4me3 binding protein, transferaseChain: A: PDB Molecule: jmjc domain-containing histone demethylation protein 1d;PDBTitle: structure of kiaa1718 jumonji domain in complex with alpha-2 ketoglutarate
15 d1j5ua_
16.4
11
Fold: MTH1598-likeSuperfamily: MTH1598-likeFamily: MTH1598-like16 c2w3pB_
15.7
14
PDB header: lyaseChain: B: PDB Molecule: benzoyl-coa-dihydrodiol lyase;PDBTitle: boxc crystal structure
17 d1jw3a_
14.2
26
Fold: MTH1598-likeSuperfamily: MTH1598-likeFamily: MTH1598-like18 d1gp6a_
12.9
12
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Penicillin synthase-like19 d1odma_
12.4
14
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Penicillin synthase-like20 c2btwA_
12.3
18
PDB header: transferaseChain: A: PDB Molecule: alr0975 protein;PDBTitle: crystal structure of alr0975
21 c2ra9A_
not modelled
9.1
10
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein duf1285;PDBTitle: crystal structure of a duf1285 family protein (sbal_2486) from2 shewanella baltica os155 at 1.40 a resolution
22 d2bu3a1
not modelled
8.4
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Phytochelatin synthase23 c2kouA_
not modelled
7.8
26
PDB header: hydrolaseChain: A: PDB Molecule: dicer-like protein 4;PDBTitle: dicer like protein
24 c1gn4B_
not modelled
7.5
10
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase;PDBTitle: h145e mutant of mycobacterium tuberculosis iron-superoxide2 dismutase.
25 c1fcuA_
not modelled
7.4
9
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronoglucosaminidase;PDBTitle: crystal structure (trigonal) of bee venom hyaluronidase
26 c3kv4A_
not modelled
7.1
20
PDB header: h3k4me3 binding protein, transferaseChain: A: PDB Molecule: phd finger protein 8;PDBTitle: structure of phf8 in complex with histone h3
27 c2i0tB_
not modelled
7.0
23
PDB header: oxidoreductaseChain: B: PDB Molecule: aromatic amine dehydrogenase;PDBTitle: crystal structure of phenylacetaldehyde derived r-2 carbinolamine adduct of aromatic amine dehydrogenase
28 c2yz3B_
not modelled
7.0
13
PDB header: hydrolaseChain: B: PDB Molecule: metallo-beta-lactamase;PDBTitle: crystallographic investigation of inhibition mode of the2 vim-2 metallo-beta-lactamase from pseudomonas aeruginosa3 with mercaptocarboxylate inhibitor
29 c3euhF_
not modelled
7.0
23
PDB header: cell cycleChain: F: PDB Molecule: muke;PDBTitle: crystal structure of the muke-mukf complex
30 c2oqkA_
not modelled
6.8
20
PDB header: translationChain: A: PDB Molecule: putative translation initiation factor eif-1a;PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
31 d1y67a2
not modelled
6.7
10
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain32 d1fcqa_
not modelled
6.4
9
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Bee venom hyaluronidase33 d1wi9a_
not modelled
6.3
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PCI domain (PINT motif)34 d1f28a_
not modelled
5.9
11
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase35 d1a9xa5
not modelled
5.9
19
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like36 c2e1nA_
not modelled
5.6
14
PDB header: circadian clock proteinChain: A: PDB Molecule: pex;PDBTitle: crystal structure of the cyanobacterium circadian clock modifier pex
37 d1kzyc2
not modelled
5.6
14
Fold: BRCT domainSuperfamily: BRCT domainFamily: 53BP138 c1ibtC_
not modelled
5.6
50
PDB header: lyaseChain: C: PDB Molecule: histidine decarboxylase beta chain;PDBTitle: structure of the d53,54n mutant of histidine decarboxylase at-170 c
39 d1uerc2
not modelled
5.6
24
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain40 d1b06a2
not modelled
5.5
19
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain41 c1kkcB_
not modelled
5.3
10
PDB header: oxidoreductaseChain: B: PDB Molecule: manganese superoxide dismutase;PDBTitle: crystal structure of aspergillus fumigatus mnsod
42 c3pu3A_
not modelled
5.2
17
PDB header: protein bindingChain: A: PDB Molecule: phd finger protein 2;PDBTitle: phf2 jumonji domain-nog complex
43 c3da4B_
not modelled
5.1
20
PDB header: antibioticChain: B: PDB Molecule: colicin-m;PDBTitle: crystal structure of colicin m, a novel phosphatase2 specifically imported by escherichia coli
44 d1gv3a2
not modelled
5.1
19
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain