| 1 |
|
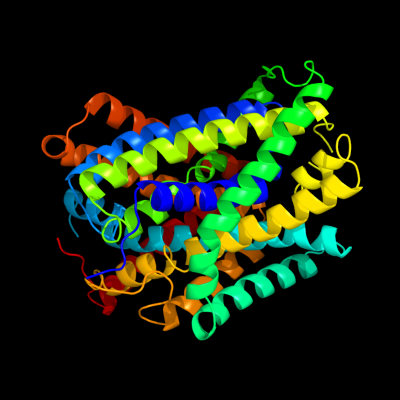
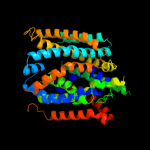
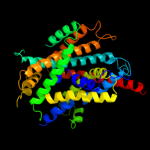
PDB 3gia chain A
Region: 5 - 439
Aligned: 423
Modelled: 423
Confidence: 100.0%
Identity: 16%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
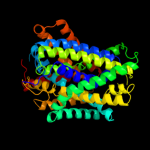
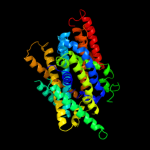
PDB 3lrc chain C
Region: 5 - 435
Aligned: 409
Modelled: 409
Confidence: 100.0%
Identity: 33%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
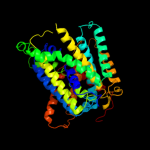
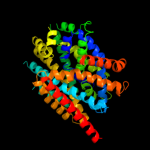
PDB 2jln chain A
Region: 2 - 439
Aligned: 425
Modelled: 427
Confidence: 100.0%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
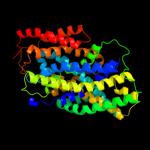
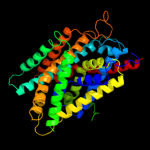
PDB 2xq2 chain A
Region: 7 - 435
Aligned: 425
Modelled: 425
Confidence: 99.4%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 5 |
|
PDB 3dh4 chain A
Region: 7 - 438
Aligned: 421
Modelled: 421
Confidence: 99.4%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 10 - 436
Aligned: 418
Modelled: 418
Confidence: 98.5%
Identity: 13%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 2 - 379
Aligned: 366
Modelled: 376
Confidence: 97.6%
Identity: 13%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 4 - 381
Aligned: 369
Modelled: 369
Confidence: 83.2%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 2l2t chain A
Region: 411 - 437
Aligned: 27
Modelled: 27
Confidence: 8.6%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 10 |
|
PDB 1lfb chain A
Region: 423 - 438
Aligned: 16
Modelled: 16
Confidence: 8.4%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Homeodomain
Phyre2
| 11 |
|
PDB 1r3j chain C
Region: 349 - 438
Aligned: 90
Modelled: 90
Confidence: 8.4%
Identity: 19%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 12 |
|
PDB 2gli chain A domain 5
Region: 429 - 439
Aligned: 11
Modelled: 11
Confidence: 8.2%
Identity: 45%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 13 |
|
PDB 2iub chain A domain 2
Region: 383 - 432
Aligned: 50
Modelled: 50
Confidence: 7.8%
Identity: 2%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 14 |
|
PDB 2rdd chain B
Region: 417 - 437
Aligned: 21
Modelled: 21
Confidence: 6.6%
Identity: 14%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 15 |
|
PDB 3rko chain F
Region: 319 - 438
Aligned: 120
Modelled: 120
Confidence: 6.5%
Identity: 9%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 16 |
|
PDB 2klu chain A
Region: 413 - 436
Aligned: 24
Modelled: 24
Confidence: 6.0%
Identity: 17%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
Phyre2
| 17 |
|
PDB 1fft chain B domain 2
Region: 344 - 411
Aligned: 68
Modelled: 68
Confidence: 5.9%
Identity: 9%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 18 |
|
PDB 3og0 chain 2
Region: 431 - 439
Aligned: 9
Modelled: 9
Confidence: 5.9%
Identity: 22%
PDB header:ribosome
Chain: 2: PDB Molecule:50s ribosomal protein l34;
PDBTitle: crystal structure of the e. coli ribosome bound to clindamycin. this2 file contains the 50s subunit of the second 70s ribosome.
Phyre2
| 19 |
|
PDB 1vt2 chain 2
Region: 431 - 439
Aligned: 9
Modelled: 9
Confidence: 5.9%
Identity: 22%
PDB header:ribosome
Chain: 2: PDB Molecule:50s ribosomal protein l34;
PDBTitle: crystal structure of the e. coli ribosome bound to cem-101. this file2 contains the 50s subunit of the second 70s ribosome.
Phyre2
| 20 |
|
PDB 3ofd chain 2
Region: 431 - 439
Aligned: 9
Modelled: 9
Confidence: 5.9%
Identity: 22%
PDB header:ribosome
Chain: 2: PDB Molecule:50s ribosomal protein l34;
PDBTitle: crystal structure of the e. coli ribosome bound to chloramphenicol.2 this file contains the 50s subunit of the second 70s ribosome.
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|