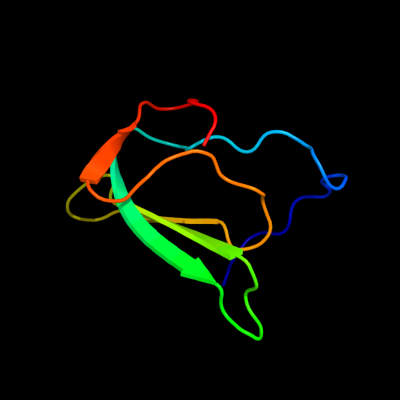
1 c2gu1A_
49.6
20
PDB header: hydrolaseChain: A: PDB Molecule: zinc peptidase;PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
2 c3k8rA_
48.0
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function (yp_427503.1) from2 rhodospirillum rubrum atcc 11170 at 2.75 a resolution
3 d1q46a2
47.5
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like4 d1e32a1
39.5
25
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like5 c2dk6A_
39.3
33
PDB header: signaling proteinChain: A: PDB Molecule: parp11 protein;PDBTitle: solution structure of wwe domain in poly (adp-ribose)2 polymerase family, member 11 (parp 11)
6 d1k75a_
35.1
14
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: L-histidinol dehydrogenase HisD7 c2hsiB_
33.7
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative peptidase m23;PDBTitle: crystal structure of putative peptidase m23 from2 pseudomonas aeruginosa, new york structural genomics3 consortium
8 c2x8nA_
26.2
25
PDB header: structural genomicsChain: A: PDB Molecule: cv0863;PDBTitle: solution nmr structure of uncharacterized protein cv08632 from chromobacterium violaceum. northeast structural3 genomics target (nesg) target cvt3. ocsp4 target cv0863.
9 c1q46A_
26.1
15
PDB header: translationChain: A: PDB Molecule: translation initiation factor 2 alpha subunit;PDBTitle: crystal structure of the eif2 alpha subunit from2 saccharomyces cerevisia
10 d2d8xa2
17.1
38
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain11 c1yc0I_
16.0
45
PDB header: hydrolase/inhibitorChain: I: PDB Molecule: kunitz-type protease inhibitor 1;PDBTitle: short form hgfa with first kunitz domain from hai-1
12 c1yz6A_
15.5
16
PDB header: translationChain: A: PDB Molecule: probable translation initiation factor 2 alphaPDBTitle: crystal structure of intact alpha subunit of aif2 from2 pyrococcus abyssi
13 d1dema_
15.0
18
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins14 d1faki_
14.9
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins15 c2kkuA_
14.2
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of protein af2351 from archaeoglobus2 fulgidus. northeast structural genomics consortium target3 att9/ontario center for structural proteomics target af2351
16 d1tfxc_
13.8
18
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins17 d1bika1
13.7
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins18 d1bika2
13.2
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins19 d1brbi_
13.0
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins20 d1bpia_
12.7
18
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins21 d1w36b3
not modelled
12.5
20
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Exodeoxyribonuclease V beta chain (RecB), C-terminal domain22 d1dtxa_
not modelled
12.2
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins23 c3bpjD_
not modelled
12.0
24
PDB header: translationChain: D: PDB Molecule: eukaryotic translation initiation factor 3 subunit j;PDBTitle: crystal structure of human translation initiation factor 3, subunit 12 alpha
24 c1y62E_
not modelled
11.7
30
PDB header: toxinChain: E: PDB Molecule: conkunitzin-s1;PDBTitle: a 2.4 crystal structure of conkunitzin-s1, a novel kunitz-2 fold cone snail neurotoxin.
25 d1irha_
not modelled
10.5
30
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins26 d1dtka_
not modelled
10.5
18
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins27 d1shpa_
not modelled
10.4
29
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins28 d1uuba_
not modelled
10.1
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins29 d1p2ji_
not modelled
10.1
18
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins30 d1go3e1
not modelled
10.0
11
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like31 c1tocR_
not modelled
10.0
30
PDB header: complex (hydrolase/inhibitor)Chain: R: PDB Molecule: ornithodorin;PDBTitle: structure of serine proteinase
32 d2brfa1
not modelled
9.8
22
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain33 d2j7ja3
not modelled
9.8
36
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H234 d1bunb_
not modelled
9.7
30
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins35 d1jc6a_
not modelled
9.1
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins36 d2z0sa1
not modelled
9.1
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like37 d1n7oa3
not modelled
9.0
60
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hyaluronate lyase-like, central domain38 d1g6xa_
not modelled
8.7
18
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins39 c3bybB_
not modelled
8.7
18
PDB header: hydrolase inhibitorChain: B: PDB Molecule: textilinin;PDBTitle: crystal structure of textilinin-1, a kunitz-type serine2 protease inhibitor from the australian common brown snake3 venom
40 d1hn0a4
not modelled
8.6
40
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hyaluronate lyase-like, central domain41 d1aapa_
not modelled
8.6
40
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins42 c3hizB_
not modelled
8.5
22
PDB header: transferase/oncoproteinChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunitPDBTitle: crystal structure of p110alpha h1047r mutant in complex with2 nish2 of p85alpha
43 d1f1sa4
not modelled
8.3
40
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hyaluronate lyase-like, central domain44 d2ahob2
not modelled
8.3
9
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like45 d1ujra_
not modelled
8.2
23
Fold: WWE domainSuperfamily: WWE domainFamily: WWE domain46 d1x1ia3
not modelled
8.1
60
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hyaluronate lyase-like, central domain47 d3ctka1
not modelled
7.9
24
Fold: Ribosome inactivating proteins (RIP)Superfamily: Ribosome inactivating proteins (RIP)Family: Plant cytotoxins48 d1zr0b1
not modelled
7.8
40
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins49 d1wjia_
not modelled
7.7
41
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain50 c3kyeC_
not modelled
7.7
26
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: roadblock/lc7 domain, robl_lc7;PDBTitle: crystal structure of roadblock/lc7 domain from streptomyces2 avermitilis
51 d1rwha3
not modelled
7.7
60
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hyaluronate lyase-like, central domain52 d1cb8a3
not modelled
7.6
60
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hyaluronate lyase-like, central domain53 d1ld6a_
not modelled
7.6
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins54 d1ejmb_
not modelled
7.5
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins55 c2ba1B_
not modelled
7.5
15
PDB header: rna binding proteinChain: B: PDB Molecule: archaeal exosome rna binding protein csl4;PDBTitle: archaeal exosome core
56 d1di2a_
not modelled
7.4
11
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)57 d1adza_
not modelled
7.4
20
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins58 d2b2ca1
not modelled
7.3
15
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Spermidine synthase59 d1ktha_
not modelled
7.1
40
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins60 c3tekA_
not modelled
7.0
26
PDB header: dna binding proteinChain: A: PDB Molecule: thermodbp-single stranded dna binding protein;PDBTitle: thermodbp: a non-canonical single-stranded dna binding protein with a2 novel structure and mechanism
61 c2l3iA_
not modelled
7.0
100
PDB header: antimicrobial proteinChain: A: PDB Molecule: aoxki4a, antimicrobial peptide in spider venom;PDBTitle: oxki4a, spider derived antimicrobial peptide
62 d2ba0a1
not modelled
6.8
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like63 d1f2fa_
not modelled
6.6
22
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain64 d1bf0a_
not modelled
6.6
10
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Small Kunitz-type inhibitors & BPTI-like toxins65 c2kcrA_
not modelled
6.6
30
PDB header: toxinChain: A: PDB Molecule: anntoxin;PDBTitle: solution structure of anntoxin
66 d1kl9a2
not modelled
6.6
13
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like67 c3tr9A_
not modelled
6.6
10
PDB header: transferaseChain: A: PDB Molecule: dihydropteroate synthase;PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
68 c2ddiA_
not modelled
6.5
30
PDB header: protein bindingChain: A: PDB Molecule: wap, follistatin/kazal, immunoglobulin, kunitzPDBTitle: nmr structure of the second kunitz domain of human wfikkn1
69 d1sdda2
not modelled
6.3
50
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins70 d2je6i1
not modelled
6.2
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like71 c2odyF_
not modelled
6.1
27
PDB header: blood clotting/blood clotting inhibitorChain: F: PDB Molecule: boophilin;PDBTitle: thrombin-bound boophilin displays a functional and accessible2 reactive-site loop
72 d1bz7a1
not modelled
6.1
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)73 d1mnda2
not modelled
5.9
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins74 c1q8kA_
not modelled
5.9
11
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 2PDBTitle: solution structure of alpha subunit of human eif2
75 c2jufA_
not modelled
5.8
19
PDB header: gene regulationChain: A: PDB Molecule: p53-associated parkin-like cytoplasmic protein;PDBTitle: nmr solution structure of parc cph domain. nesg target2 hr3443b/sgc-toronto
76 d2eyva1
not modelled
5.5
22
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain77 c2jo7A_
not modelled
5.4
24
PDB header: surface active proteinChain: A: PDB Molecule: glycosylphosphatidylinositol-anchored merozoitePDBTitle: solution structure of the adhesion protein bd37 from2 babesia divergens
78 c1facA_
not modelled
5.4
50
PDB header: coagulation factorChain: A: PDB Molecule: coagulation factor viii;PDBTitle: coagulation factor viii, nmr, 1 structure
79 d1qe3a_
not modelled
5.4
33
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like80 c3ayhA_
not modelled
5.3
12
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase iii subunit rpc9;PDBTitle: crystal structure of the c17/25 subcomplex from s. pombe rna2 polymerase iii
81 d1d0xa2
not modelled
5.2
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins82 d2i27n1
not modelled
5.2
11
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)83 d1il1b1
not modelled
5.1
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)84 c2kz5A_
not modelled
5.1
25
PDB header: transcriptionChain: A: PDB Molecule: transcription factor nf-e2 45 kda subunit;PDBTitle: solution nmr structure of transcription factor nf-e2 subunit's dna2 binding domain from homo sapiens, northeast structural genomics3 consortium target hr4653b
85 d1ouoa_
not modelled
5.0
14
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: Endonuclease I