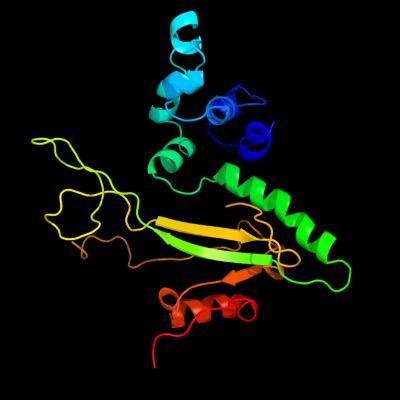
1 d1musa_
98.6
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)2 d1b7ea_
97.3
10
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)3 d1yoca1
49.5
22
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: PaaI/YdiI-like4 d1wb9a2
46.7
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like5 c3thxB_
32.6
18
PDB header: dna binding protein/dnaChain: B: PDB Molecule: dna mismatch repair protein msh3;PDBTitle: human mutsbeta complexed with an idl of 3 bases (loop3) and adp
6 c1d5yD_
26.2
20
PDB header: transcription/dnaChain: D: PDB Molecule: rob transcription factor;PDBTitle: crystal structure of the e. coli rob transcription factor2 in complex with dna
7 c1ewqA_
20.7
21
PDB header: replication/dnaChain: A: PDB Molecule: dna mismatch repair protein muts;PDBTitle: crystal structure taq muts complexed with a heteroduplex2 dna at 2.2 a resolution
8 c1wbdA_
16.6
27
PDB header: dna-bindingChain: A: PDB Molecule: dna mismatch repair protein muts;PDBTitle: crystal structure of e. coli dna mismatch repair enzyme2 muts, e38q mutant, in complex with a g.t mismatch
9 d1u35c1
12.2
17
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones10 c2o8bA_
12.0
18
PDB header: dna binding protein/dnaChain: A: PDB Molecule: dna mismatch repair protein msh2;PDBTitle: human mutsalpha (msh2/msh6) bound to adp and a g t mispair
11 c2o8dB_
11.6
21
PDB header: dna binding protein/dnaChain: B: PDB Molecule: dna mismatch repair protein msh6;PDBTitle: human mutsalpha (msh2/msh6) bound to adp and a g du mispair
12 c3pcoD_
11.2
26
PDB header: ligaseChain: D: PDB Molecule: phenylalanyl-trna synthetase, beta chain;PDBTitle: crystal structure of e. coli phenylalanine-trna synthetase complexed2 with phenylalanine and amp
13 d2jfga2
10.7
14
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain14 c2f8nK_
8.9
17
PDB header: structural protein/dnaChain: K: PDB Molecule: histone h2a type 1;PDBTitle: 2.9 angstrom x-ray structure of hybrid macroh2a nucleosomes
15 c2hmhA_
8.8
11
PDB header: cytokine regulatorChain: A: PDB Molecule: suppressor of cytokine signaling 3;PDBTitle: crystal structure of socs3 in complex with gp130(ptyr757)2 phosphopeptide.
16 c3czpA_
8.8
26
PDB header: transferaseChain: A: PDB Molecule: putative polyphosphate kinase 2;PDBTitle: crystal structure of putative polyphosphate kinase 2 from pseudomonas2 aeruginosa pa01
17 d1k8ke_
8.8
24
Fold: Arp2/3 complex 21 kDa subunit ARPC3Superfamily: Arp2/3 complex 21 kDa subunit ARPC3Family: Arp2/3 complex 21 kDa subunit ARPC318 d1ewqa2
8.4
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like19 d1id3c_
7.7
13
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones20 d1eqza_
7.7
15
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones21 c3btpA_
not modelled
7.6
20
PDB header: dna binding protein, chaperoneChain: A: PDB Molecule: single-strand dna-binding protein;PDBTitle: crystal structure of agrobacterium tumefaciens vire2 in complex with2 its chaperone vire1: a novel fold and implications for dna binding
22 c3tdqB_
not modelled
7.6
36
PDB header: cell adhesionChain: B: PDB Molecule: pily2 protein;PDBTitle: crystal structure of a fimbrial biogenesis protein pily22 (pily2_pa4555) from pseudomonas aeruginosa pao1 at 2.10 a resolution
23 c2vzaD_
not modelled
7.4
18
PDB header: cell adhesionChain: D: PDB Molecule: cell filamentation protein;PDBTitle: type iv secretion system effector protein bepa
24 d1tzya_
not modelled
7.0
15
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones25 d1dk7a_
not modelled
6.9
43
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain26 d1sjpa2
not modelled
6.6
57
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain27 c3rhfB_
not modelled
6.5
21
PDB header: transferaseChain: B: PDB Molecule: putative polyphosphate kinase 2 family protein;PDBTitle: crystal structure of polyphosphate kinase 2 from arthrobacter2 aurescens tc1
28 d1srva_
not modelled
6.4
57
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain29 d1ioka2
not modelled
6.2
43
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain30 d1aoic_
not modelled
6.1
19
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones31 d1ikpa3
not modelled
6.0
69
Fold: Toxins' membrane translocation domainsSuperfamily: Exotoxin A, middle domainFamily: Exotoxin A, middle domain32 c2vifA_
not modelled
6.0
23
PDB header: signaling proteinChain: A: PDB Molecule: suppressor of cytokine signalling 6;PDBTitle: crystal structure of socs6 sh2 domain in complex with a c-kit2 phosphopeptide
33 c3ld9D_
not modelled
5.9
13
PDB header: transferaseChain: D: PDB Molecule: thymidylate kinase;PDBTitle: crystal structure of thymidylate kinase from ehrlichia chaffeensis at2 2.15a resolution
34 d1oela2
not modelled
5.7
43
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain35 d1kx3c_
not modelled
5.7
19
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones36 c3pjvD_
not modelled
5.5
50
PDB header: lyaseChain: D: PDB Molecule: cyclic dimeric gmp binding protein;PDBTitle: structure of pseudomonas fluorescence lapd periplasmic domain
37 d1pvda3
not modelled
5.5
2
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module38 d1sjpa3
not modelled
5.3
0
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: GroEL-like chaperone, intermediate domain