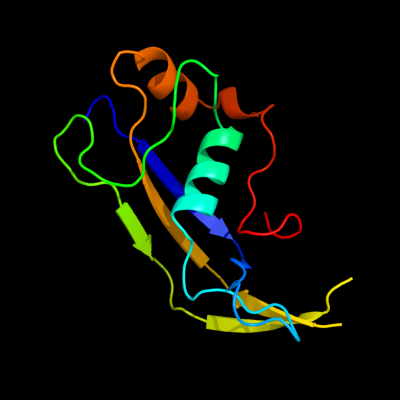
1 c3kg4A_
99.7
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from mannheimia2 succiniciproducens
2 d2ihta1
64.8
26
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain3 d1ovma1
63.5
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain4 d1t9ba1
52.7
21
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain5 d2ez9a1
51.5
20
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain6 d1zpda1
51.2
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain7 d1pvda1
49.7
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain8 d2djia1
41.4
15
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain9 d2ji7a1
35.2
15
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain10 d1fyva_
29.3
17
Fold: Flavodoxin-likeSuperfamily: Toll/Interleukin receptor TIR domainFamily: Toll/Interleukin receptor TIR domain11 c3h16A_
26.2
7
PDB header: signaling proteinChain: A: PDB Molecule: tir protein;PDBTitle: crystal structure of a bacteria tir domain, pdtir from2 paracoccus denitrificans
12 d1ozha1
24.9
8
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain13 d1q6za1
23.9
26
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain14 c3oziB_
22.9
8
PDB header: plant proteinChain: B: PDB Molecule: l6tr;PDBTitle: crystal structure of the tir domain from the flax disease resistance2 protein l6
15 c2vbgB_
22.1
17
PDB header: lyaseChain: B: PDB Molecule: branched-chain alpha-ketoacid decarboxylase;PDBTitle: the complex structure of the branched-chain keto acid2 decarboxylase (kdca) from lactococcus lactis with 2r-1-3 hydroxyethyl-deazathdp
16 c2panF_
18.0
18
PDB header: lyaseChain: F: PDB Molecule: glyoxylate carboligase;PDBTitle: crystal structure of e. coli glyoxylate carboligase
17 c1powA_
14.5
21
PDB header: oxidoreductase(oxygen as acceptor)Chain: A: PDB Molecule: pyruvate oxidase;PDBTitle: the refined structures of a stabilized mutant and of wild-type2 pyruvate oxidase from lactobacillus plantarum
18 c1wxnA_
12.3
50
PDB header: toxinChain: A: PDB Molecule: toxin apetx2;PDBTitle: solution structure of apetx2, a specific peptide inhibitor2 of asic3 proton-gated channels
19 c2vbiF_
11.2
15
PDB header: lyaseChain: F: PDB Molecule: pyruvate decarboxylase;PDBTitle: holostructure of pyruvate decarboxylase from acetobacter2 pasteurianus
20 d2nzwa1
11.0
14
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: FucT-like21 d1fyxa_
not modelled
10.4
4
Fold: Flavodoxin-likeSuperfamily: Toll/Interleukin receptor TIR domainFamily: Toll/Interleukin receptor TIR domain22 c2ap7A_
not modelled
10.2
60
PDB header: antibioticChain: A: PDB Molecule: bombinin h2;PDBTitle: solution structure of bombinin h2 in dpc micelles
23 c3pdiG_
not modelled
10.1
13
PDB header: protein bindingChain: G: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nife;PDBTitle: precursor bound nifen
24 c2ap8A_
not modelled
10.1
60
PDB header: antibioticChain: A: PDB Molecule: bombinin h4;PDBTitle: solution structure of bombinin h4 in dpc micelles
25 c2w93A_
not modelled
9.6
17
PDB header: lyaseChain: A: PDB Molecule: pyruvate decarboxylase isozyme 1;PDBTitle: crystal structure of the saccharomyces cerevisiae pyruvate2 decarboxylase variant e477q in complex with the surrogate3 pyruvamide
26 c2djiA_
not modelled
9.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate oxidase;PDBTitle: crystal structure of pyruvate oxidase from aerococcus2 viridans containing fad
27 c2z5vA_
not modelled
8.6
16
PDB header: immune systemChain: A: PDB Molecule: myeloid differentiation primary response proteinPDBTitle: solution structure of the tir domain of human myd88
28 c2pgnA_
not modelled
7.7
24
PDB header: hydrolaseChain: A: PDB Molecule: cyclohexane-1,2-dione hydrolase (cdh);PDBTitle: the crystal structure of fad and thdp-dependent cyclohexane-1,2-dione2 hydrolase in complex with cyclohexane-1,2-dione
29 d1m1na_
not modelled
7.6
7
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein30 d2fug61
not modelled
7.1
17
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nq06-like31 d1vkya_
not modelled
7.0
28
Fold: QueA-likeSuperfamily: QueA-likeFamily: QueA-like32 d1qh8a_
not modelled
6.9
7
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein33 c3le4A_
not modelled
6.8
27
PDB header: nuclear proteinChain: A: PDB Molecule: microprocessor complex subunit dgcr8;PDBTitle: crystal structure of the dgcr8 dimerization domain
34 c1jscA_
not modelled
6.6
21
PDB header: lyaseChain: A: PDB Molecule: acetohydroxy-acid synthase;PDBTitle: crystal structure of the catalytic subunit of yeast2 acetohydroxyacid synthase: a target for herbicidal3 inhibitors
35 c1ovmC_
not modelled
5.9
18
PDB header: lyaseChain: C: PDB Molecule: indole-3-pyruvate decarboxylase;PDBTitle: crystal structure of indolepyruvate decarboxylase from2 enterobacter cloacae
36 d1wdia_
not modelled
5.5
38
Fold: QueA-likeSuperfamily: QueA-likeFamily: QueA-like37 c3cf4G_
not modelled
5.2
9
PDB header: oxidoreductaseChain: G: PDB Molecule: acetyl-coa decarboxylase/synthase epsilon subunit;PDBTitle: structure of the codh component of the m. barkeri acds complex