| 1 |
|
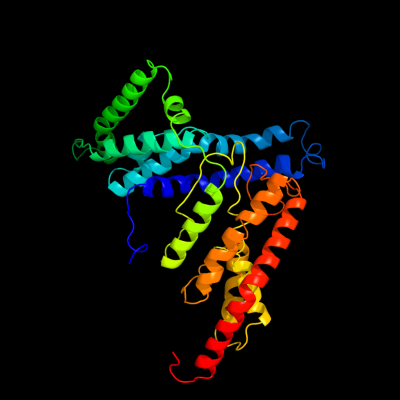
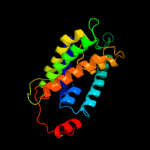
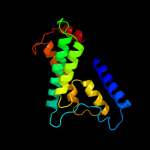
PDB 3qnq chain D
Region: 2 - 353
Aligned: 350
Modelled: 352
Confidence: 100.0%
Identity: 17%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 2 |
|
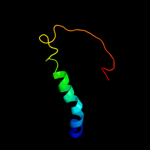
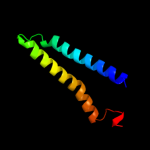
PDB 2b2h chain A
Region: 191 - 353
Aligned: 162
Modelled: 163
Confidence: 82.0%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 3 |
|
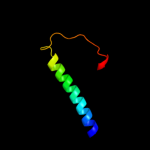
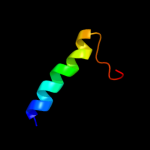
PDB 3hd6 chain A
Region: 209 - 357
Aligned: 145
Modelled: 149
Confidence: 79.5%
Identity: 16%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 4 |
|
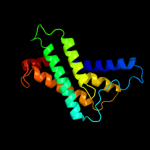
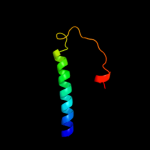
PDB 2knc chain A
Region: 312 - 355
Aligned: 44
Modelled: 44
Confidence: 76.3%
Identity: 14%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 5 |
|
PDB 3b9y chain A
Region: 191 - 355
Aligned: 162
Modelled: 165
Confidence: 66.2%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 6 |
|
PDB 2jln chain A
Region: 321 - 357
Aligned: 37
Modelled: 37
Confidence: 42.6%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 7 |
|
PDB 1j4n chain A
Region: 103 - 352
Aligned: 222
Modelled: 222
Confidence: 42.2%
Identity: 10%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 8 |
|
PDB 2oar chain A domain 1
Region: 312 - 353
Aligned: 42
Modelled: 42
Confidence: 26.6%
Identity: 2%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 9 |
|
PDB 3d9s chain B
Region: 137 - 357
Aligned: 193
Modelled: 197
Confidence: 18.2%
Identity: 15%
PDB header:membrane protein
Chain: B: PDB Molecule:aquaporin-5;
PDBTitle: human aquaporin 5 (aqp5) - high resolution x-ray structure
Phyre2
| 10 |
|
PDB 1fft chain B domain 2
Region: 313 - 358
Aligned: 46
Modelled: 46
Confidence: 14.6%
Identity: 9%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 11 |
|
PDB 3dtu chain B domain 2
Region: 312 - 355
Aligned: 44
Modelled: 44
Confidence: 13.5%
Identity: 11%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 12 |
|
PDB 1ymg chain A
Region: 177 - 353
Aligned: 149
Modelled: 152
Confidence: 13.2%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
Phyre2
| 13 |
|
PDB 1ymg chain A domain 1
Region: 177 - 353
Aligned: 149
Modelled: 152
Confidence: 13.2%
Identity: 14%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 14 |
|
PDB 3ehb chain B domain 2
Region: 312 - 355
Aligned: 44
Modelled: 44
Confidence: 11.0%
Identity: 9%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 15 |
|
PDB 2oar chain A
Region: 279 - 353
Aligned: 75
Modelled: 75
Confidence: 7.1%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 16 |
|
PDB 2ket chain A
Region: 2 - 26
Aligned: 21
Modelled: 25
Confidence: 6.9%
Identity: 24%
PDB header:antibiotic
Chain: A: PDB Molecule:cathelicidin-6;
PDBTitle: solution structure of bmap-27
Phyre2
| 17 |
|
PDB 2ht2 chain B
Region: 108 - 351
Aligned: 228
Modelled: 229
Confidence: 6.1%
Identity: 9%
PDB header:membrane protein
Chain: B: PDB Molecule:h(+)/cl(-) exchange transporter clca;
PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
Phyre2
| 18 |
|
PDB 1qle chain B
Region: 312 - 356
Aligned: 45
Modelled: 45
Confidence: 5.9%
Identity: 9%
PDB header:oxidoreductase/immune system
Chain: B: PDB Molecule:cytochrome c oxidase polypeptide ii;
PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
Phyre2