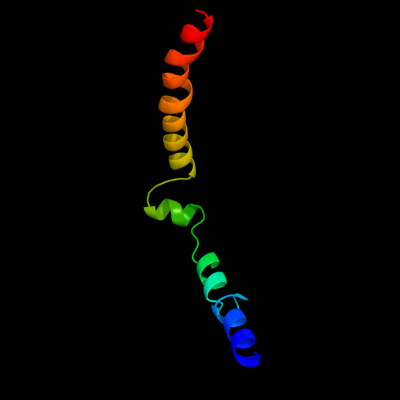
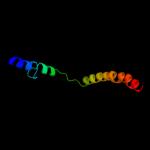
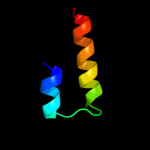
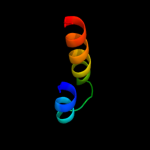
1 d1rhzb_
51.6
14
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit2 d1r0ka1
19.4
16
Fold: Left-handed superhelixSuperfamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domainFamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain3 d1q0qa1
19.2
8
Fold: Left-handed superhelixSuperfamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domainFamily: 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain4 c2y0sQ_
18.6
17
PDB header: transferaseChain: Q: PDB Molecule: rna polymerase subunit 13;PDBTitle: crystal structure of sulfolobus shibatae rna polymerase in2 p21 space group
5 c2y0sJ_
18.6
17
PDB header: transferaseChain: J: PDB Molecule: rna polymerase subunit 13;PDBTitle: crystal structure of sulfolobus shibatae rna polymerase in2 p21 space group
6 d1smya1
14.5
20
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RNA polymerase alpha subunit dimerisation domain7 d2j5pa1
14.3
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FtsK C-terminal domain-like8 c2jcyA_
13.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: x-ray structure of mutant 1-deoxy-d-xylulose 5-phosphate2 reductoisomerase, dxr, rv2870c, from mycobacterium3 tuberculosis
9 d2ve8a1
13.0
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FtsK C-terminal domain-like10 c2kxaA_
12.9
50
PDB header: viral protein, immune systemChain: A: PDB Molecule: haemagglutinin ha2 chain peptide;PDBTitle: the hemagglutinin fusion peptide (h1 subtype) at ph 7.4
11 c2vayB_
12.1
45
PDB header: metal transportChain: B: PDB Molecule: voltage-dependent l-type calcium channel subunitPDBTitle: calmodulin complexed with cav1.1 iq peptide
12 c2wb1Q_
11.5
28
PDB header: transcriptionChain: Q: PDB Molecule: dna-directed rna polymerase rpo13 subunit;PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
13 c2k6lA_
10.7
33
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the solution structure of xacb0070 from xanthonomas2 axonopodis pv citri reveals this new protein is a member of3 the rhh family of transcriptional repressors
14 c2wb1J_
10.5
20
PDB header: transcriptionChain: J: PDB Molecule: dna-directed rna polymerase rpo13 subunit;PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
15 d1rh5b_
8.9
42
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit16 c2bo9B_
8.2
19
PDB header: hydrolaseChain: B: PDB Molecule: human latexin;PDBTitle: human carboxypeptidase a4 in complex with human latexin.
17 c2fmlB_
8.1
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: mutt/nudix family protein;PDBTitle: crystal structure of mutt/nudix family protein from enterococcus2 faecalis
18 d1ynja1
7.9
18
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RNA polymerase alpha subunit dimerisation domain19 c3gz6A_
7.7
21
PDB header: dna binding protein/dnaChain: A: PDB Molecule: mutt/nudix family protein;PDBTitle: crystal structure of shewanella oneidensis nrtr complexed2 with a 27mer dna
20 c2wwbB_
7.5
14
PDB header: ribosomeChain: B: PDB Molecule: protein transport protein sec61 subunit gamma;PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
21 c2zykA_
not modelled
6.7
15
PDB header: sugar binding proteinChain: A: PDB Molecule: solute-binding protein;PDBTitle: crystal structure of cyclo/maltodextrin-binding protein2 complexed with gamma-cyclodextrin
22 d1bdfa1
not modelled
6.6
26
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RNA polymerase alpha subunit dimerisation domain23 c2pd0D_
not modelled
6.6
44
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: protein cgd2_2020 from cryptosporidium parvum
24 d2fb1a1
not modelled
6.5
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Nudix-associated domain25 c1r0lD_
not modelled
6.3
16
PDB header: oxidoreductaseChain: D: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from2 zymomonas mobilis in complex with nadph
26 c3uorB_
not modelled
5.8
13
PDB header: sugar binding proteinChain: B: PDB Molecule: abc transporter sugar binding protein;PDBTitle: the structure of the sugar-binding protein male from the phytopathogen2 xanthomonas citri
27 c3hkzZ_
not modelled
5.6
20
PDB header: transferaseChain: Z: PDB Molecule: dna-directed rna polymerase subunit 13;PDBTitle: the x-ray crystal structure of rna polymerase from archaea
28 c2uvgA_
not modelled
5.6
24
PDB header: sugar-binding proteinChain: A: PDB Molecule: abc type periplasmic sugar-binding protein;PDBTitle: structure of a periplasmic oligogalacturonide binding2 protein from yersinia enterocolitica
29 c3kk4B_
not modelled
5.5
32
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein bp1543;PDBTitle: uncharacterized protein bp1543 from bordetella pertussis tohama i
30 c3a14B_
not modelled
5.5
16
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
31 c3au9A_
not modelled
5.4
22
PDB header: isomerase/isomerase inhibitorChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of the quaternary complex-1 of an isomerase
32 c2eghA_
not modelled
5.4
8
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase2 complexed with a magnesium ion, nadph and fosmidomycin
33 c2fb1A_
not modelled
5.4
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of protein bt0354 from bacteroides thetaiotaomicron
34 d1jb0i_
not modelled
5.3
30
Fold: Single transmembrane helixSuperfamily: Subunit VIII of photosystem I reaction centre, PsaIFamily: Subunit VIII of photosystem I reaction centre, PsaI