| 1 |
|
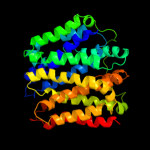
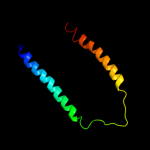
PDB 1pw4 chain A
Region: 1 - 405
Aligned: 399
Modelled: 400
Confidence: 100.0%
Identity: 13%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
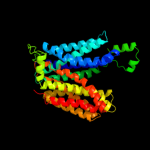
PDB 1pv7 chain A
Region: 12 - 402
Aligned: 386
Modelled: 391
Confidence: 100.0%
Identity: 14%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 3 |
|
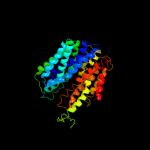
PDB 2gfp chain A
Region: 16 - 391
Aligned: 368
Modelled: 376
Confidence: 99.9%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 4 |
|
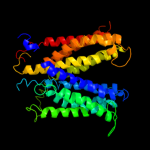
PDB 3o7p chain A
Region: 14 - 400
Aligned: 380
Modelled: 387
Confidence: 99.9%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 18 - 394
Aligned: 377
Modelled: 377
Confidence: 99.9%
Identity: 11%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3mku chain A
Region: 6 - 407
Aligned: 402
Modelled: 402
Confidence: 37.7%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:multi antimicrobial extrusion protein (na(+)/drug
PDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
Phyre2
| 7 |
|
PDB 3lrc chain C
Region: 277 - 404
Aligned: 123
Modelled: 128
Confidence: 28.3%
Identity: 11%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 8 |
|
PDB 3rko chain F
Region: 375 - 406
Aligned: 32
Modelled: 32
Confidence: 22.7%
Identity: 16%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 9 |
|
PDB 2g9p chain A
Region: 73 - 86
Aligned: 14
Modelled: 14
Confidence: 20.9%
Identity: 36%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 10 |
|
PDB 2bbj chain B
Region: 336 - 402
Aligned: 65
Modelled: 67
Confidence: 16.9%
Identity: 17%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 11 |
|
PDB 2k9p chain A
Region: 368 - 404
Aligned: 37
Modelled: 37
Confidence: 13.6%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:pheromone alpha factor receptor;
PDBTitle: structure of tm1_tm2 in lppg micelles
Phyre2
| 12 |
|
PDB 2bbn chain B
Region: 10 - 23
Aligned: 14
Modelled: 14
Confidence: 9.9%
Identity: 29%
PDB header:calcium-binding protein
Chain: B: PDB Molecule:myosin light chain kinase;
PDBTitle: solution structure of a calmodulin-target peptide complex2 by multidimensional nmr
Phyre2
| 13 |
|
PDB 2bbm chain B
Region: 10 - 23
Aligned: 14
Modelled: 14
Confidence: 9.9%
Identity: 29%
PDB header:calcium-binding protein
Chain: B: PDB Molecule:myosin light chain kinase;
PDBTitle: solution structure of a calmodulin-target peptide complex2 by multidimensional nmr
Phyre2
| 14 |
|
PDB 3bl2 chain A domain 1
Region: 10 - 39
Aligned: 30
Modelled: 30
Confidence: 9.2%
Identity: 23%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 15 |
|
PDB 3hd6 chain A
Region: 5 - 219
Aligned: 206
Modelled: 215
Confidence: 8.2%
Identity: 8%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 16 |
|
PDB 2k21 chain A
Region: 378 - 408
Aligned: 31
Modelled: 31
Confidence: 6.7%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e
PDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
Phyre2
| 17 |
|
PDB 3b60 chain A domain 2
Region: 10 - 84
Aligned: 75
Modelled: 75
Confidence: 6.6%
Identity: 17%
Fold: ABC transporter transmembrane region
Superfamily: ABC transporter transmembrane region
Family: ABC transporter transmembrane region
Phyre2
| 18 |
|
PDB 3kf9 chain D
Region: 10 - 23
Aligned: 14
Modelled: 14
Confidence: 6.4%
Identity: 29%
PDB header:cell cycle/calcium-binding protein
Chain: D: PDB Molecule:myosin light chain kinase 2, skeletal/cardiac muscle;
PDBTitle: crystal structure of the sdcen/skmlck complex
Phyre2
| 19 |
|
PDB 1pf4 chain A domain 2
Region: 10 - 84
Aligned: 75
Modelled: 75
Confidence: 6.4%
Identity: 9%
Fold: ABC transporter transmembrane region
Superfamily: ABC transporter transmembrane region
Family: ABC transporter transmembrane region
Phyre2
| 20 |
|
PDB 3kf9 chain B
Region: 10 - 23
Aligned: 14
Modelled: 14
Confidence: 6.2%
Identity: 29%
PDB header:cell cycle/calcium-binding protein
Chain: B: PDB Molecule:myosin light chain kinase 2, skeletal/cardiac muscle;
PDBTitle: crystal structure of the sdcen/skmlck complex
Phyre2
| 21 |
|