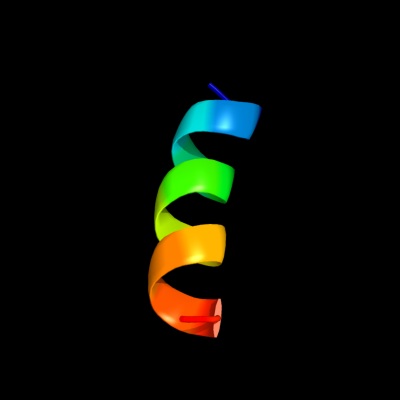| 1 |
|
PDB 3arc chain L
Region: 7 - 21
Aligned: 15
Modelled: 15
Confidence: 16.3%
Identity: 13%
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 2 |
|
PDB 2wj8 chain N
Region: 9 - 22
Aligned: 14
Modelled: 14
Confidence: 16.2%
Identity: 21%
PDB header:rna binding protein/rna
Chain: N: PDB Molecule:nucleoprotein;
PDBTitle: respiratory syncitial virus ribonucleoprotein
Phyre2
| 3 |
|
PDB 1zll chain E
Region: 3 - 24
Aligned: 22
Modelled: 22
Confidence: 15.6%
Identity: 32%
PDB header:membrane protein/signaling protein
Chain: E: PDB Molecule:cardiac phospholamban;
PDBTitle: nmr structure of unphosphorylated human phospholamban2 pentamer
Phyre2
| 4 |
|
PDB 1xme chain C domain 1
Region: 2 - 21
Aligned: 20
Modelled: 20
Confidence: 12.8%
Identity: 30%
Fold: Single transmembrane helix
Superfamily: Bacterial ba3 type cytochrome c oxidase subunit IIa
Family: Bacterial ba3 type cytochrome c oxidase subunit IIa
Phyre2
| 5 |
|
PDB 1prt chain C domain 1
Region: 28 - 51
Aligned: 24
Modelled: 24
Confidence: 10.5%
Identity: 25%
Fold: OB-fold
Superfamily: Bacterial enterotoxins
Family: Bacterial AB5 toxins, B-subunits
Phyre2
| 6 |
|
PDB 1h9m chain A domain 2
Region: 25 - 48
Aligned: 24
Modelled: 24
Confidence: 9.9%
Identity: 25%
Fold: OB-fold
Superfamily: MOP-like
Family: BiMOP, duplicated molybdate-binding domain
Phyre2
| 7 |
|
PDB 1h9r chain A domain 1
Region: 25 - 46
Aligned: 22
Modelled: 22
Confidence: 9.0%
Identity: 14%
Fold: OB-fold
Superfamily: MOP-like
Family: BiMOP, duplicated molybdate-binding domain
Phyre2
| 8 |
|
PDB 1prt chain B domain 1
Region: 28 - 51
Aligned: 24
Modelled: 24
Confidence: 8.9%
Identity: 29%
Fold: OB-fold
Superfamily: Bacterial enterotoxins
Family: Bacterial AB5 toxins, B-subunits
Phyre2
| 9 |
|
PDB 1y4o chain A domain 1
Region: 3 - 41
Aligned: 39
Modelled: 39
Confidence: 8.8%
Identity: 26%
Fold: Profilin-like
Superfamily: Roadblock/LC7 domain
Family: Roadblock/LC7 domain
Phyre2
| 10 |
|
PDB 2krx chain A
Region: 26 - 37
Aligned: 12
Modelled: 12
Confidence: 8.1%
Identity: 25%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:asl3597 protein;
PDBTitle: solution nmr structure of asl3597 from nostoc sp. pcc7120. northeast2 structural genomics consortium target id nsr244.
Phyre2
| 11 |
|
PDB 1h9m chain A domain 1
Region: 25 - 46
Aligned: 22
Modelled: 22
Confidence: 7.2%
Identity: 27%
Fold: OB-fold
Superfamily: MOP-like
Family: BiMOP, duplicated molybdate-binding domain
Phyre2
| 12 |
|
PDB 1fr3 chain A
Region: 25 - 46
Aligned: 22
Modelled: 22
Confidence: 6.9%
Identity: 18%
Fold: OB-fold
Superfamily: MOP-like
Family: Molybdate/tungstate binding protein MOP
Phyre2