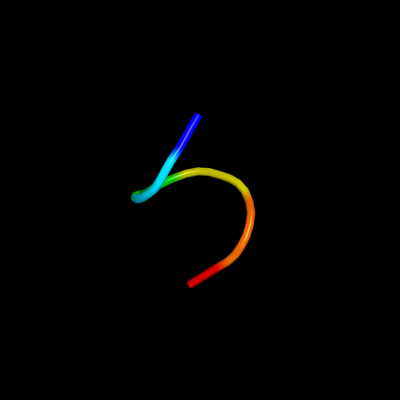| 1 |
|
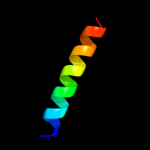
PDB 1upt chain D
Region: 180 - 188
Aligned: 9
Modelled: 6
Confidence: 19.7%
Identity: 22%
Fold: GRIP domain
Superfamily: GRIP domain
Family: GRIP domain
Phyre2
| 2 |
|
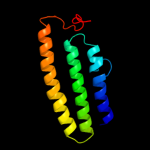
PDB 2j5d chain A
Region: 137 - 159
Aligned: 23
Modelled: 23
Confidence: 18.8%
Identity: 22%
PDB header:membrane protein
Chain: A: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: nmr structure of bnip3 transmembrane domain in lipid2 bicelles
Phyre2
| 3 |
|
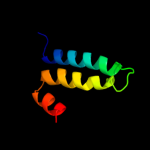
PDB 2ka2 chain B
Region: 137 - 158
Aligned: 22
Modelled: 22
Confidence: 18.0%
Identity: 23%
PDB header:membrane protein
Chain: B: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
Phyre2
| 4 |
|
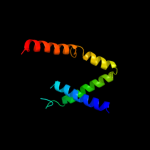
PDB 2ka1 chain A
Region: 137 - 158
Aligned: 22
Modelled: 22
Confidence: 18.0%
Identity: 23%
PDB header:membrane protein
Chain: A: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
Phyre2
| 5 |
|
PDB 2ka2 chain A
Region: 137 - 158
Aligned: 22
Modelled: 22
Confidence: 18.0%
Identity: 23%
PDB header:membrane protein
Chain: A: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
Phyre2
| 6 |
|
PDB 2ka1 chain B
Region: 137 - 158
Aligned: 22
Modelled: 22
Confidence: 18.0%
Identity: 23%
PDB header:membrane protein
Chain: B: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
Phyre2
| 7 |
|
PDB 3rko chain K
Region: 1 - 104
Aligned: 98
Modelled: 104
Confidence: 16.0%
Identity: 9%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 8 |
|
PDB 3few chain X
Region: 112 - 164
Aligned: 53
Modelled: 53
Confidence: 6.6%
Identity: 19%
PDB header:immune system
Chain: X: PDB Molecule:colicin s4;
PDBTitle: structure and function of colicin s4, a colicin with a2 duplicated receptor binding domain
Phyre2
| 9 |
|
PDB 3hfx chain A
Region: 3 - 95
Aligned: 89
Modelled: 93
Confidence: 6.4%
Identity: 6%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 10 |
|
PDB 1a87 chain A
Region: 107 - 167
Aligned: 61
Modelled: 61
Confidence: 6.1%
Identity: 15%
Fold: Toxins' membrane translocation domains
Superfamily: Colicin
Family: Colicin
Phyre2
| 11 |
|
PDB 1a87 chain A
Region: 107 - 167
Aligned: 61
Modelled: 61
Confidence: 6.1%
Identity: 15%
PDB header:bacteriocin
Chain: A: PDB Molecule:colicin n;
PDBTitle: colicin n
Phyre2