| 1 |
|
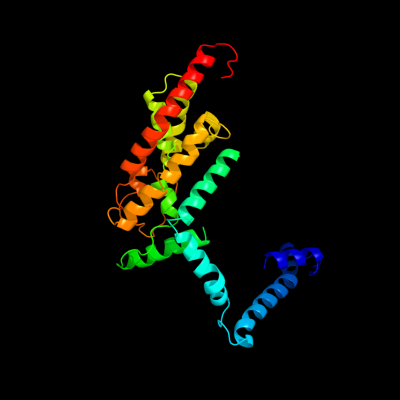
PDB 3qnq chain D
Region: 198 - 464
Aligned: 265
Modelled: 267
Confidence: 92.3%
Identity: 11%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 2 |
|
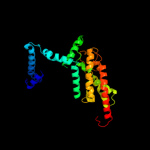
PDB 2oar chain A domain 1
Region: 371 - 463
Aligned: 93
Modelled: 93
Confidence: 90.7%
Identity: 13%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 3 |
|
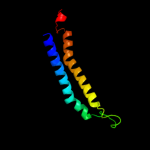
PDB 2oar chain A
Region: 371 - 463
Aligned: 93
Modelled: 93
Confidence: 89.4%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 4 |
|
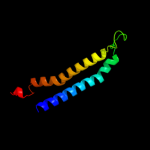
PDB 3hzq chain A
Region: 371 - 454
Aligned: 83
Modelled: 84
Confidence: 77.2%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: structure of a tetrameric mscl in an expanded intermediate2 state
Phyre2
| 5 |
|
PDB 5rub chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 15.4%
Identity: 19%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 6 |
|
PDB 1gk8 chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 14.3%
Identity: 23%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 7 |
|
PDB 1bwv chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 11.7%
Identity: 29%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 8 |
|
PDB 1rbl chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 11.4%
Identity: 23%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 9 |
|
PDB 2d69 chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 11.0%
Identity: 16%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 10 |
|
PDB 2knc chain A
Region: 406 - 461
Aligned: 54
Modelled: 56
Confidence: 10.3%
Identity: 19%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 11 |
|
PDB 8ruc chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 9.8%
Identity: 23%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 12 |
|
PDB 2bbj chain B
Region: 390 - 450
Aligned: 50
Modelled: 61
Confidence: 9.8%
Identity: 22%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 13 |
|
PDB 1bxn chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 9.6%
Identity: 19%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 14 |
|
PDB 1geh chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 9.1%
Identity: 29%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 15 |
|
PDB 1svd chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 9.1%
Identity: 16%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 16 |
|
PDB 1ots chain A
Region: 379 - 463
Aligned: 85
Modelled: 85
Confidence: 8.3%
Identity: 12%
Fold: Clc chloride channel
Superfamily: Clc chloride channel
Family: Clc chloride channel
Phyre2
| 17 |
|
PDB 1ej7 chain L domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 6.9%
Identity: 23%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 18 |
|
PDB 2zvi chain B
Region: 198 - 226
Aligned: 29
Modelled: 29
Confidence: 5.8%
Identity: 7%
PDB header:isomerase
Chain: B: PDB Molecule:2,3-diketo-5-methylthiopentyl-1-phosphate
PDBTitle: crystal structure of 2,3-diketo-5-methylthiopentyl-1-2 phosphate enolase from bacillus subtilis
Phyre2
| 19 |
|
PDB 1wdd chain A domain 2
Region: 196 - 226
Aligned: 31
Modelled: 31
Confidence: 5.5%
Identity: 23%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 20 |
|
PDB 1w2w chain J
Region: 282 - 313
Aligned: 30
Modelled: 32
Confidence: 5.5%
Identity: 30%
PDB header:isomerase
Chain: J: PDB Molecule:5-methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of yeast ypr118w, a methylthioribose-1-2 phosphate isomerase related to regulatory eif2b subunits
Phyre2